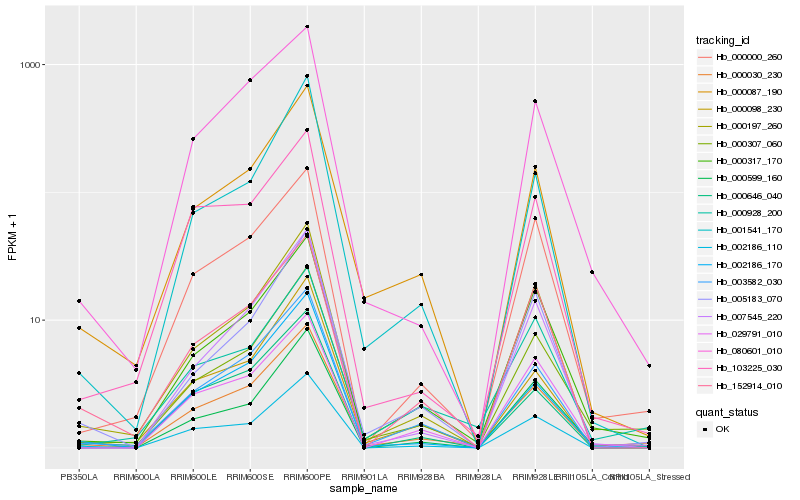
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000030_230 |
0.0 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 2 |
Hb_000307_060 |
0.0726259447 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 3 |
Hb_003582_030 |
0.0837361838 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 4 |
Hb_000317_170 |
0.08837006 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 5 |
Hb_000087_190 |
0.0944248109 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 6 |
Hb_000000_260 |
0.095171058 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 7 |
Hb_002186_110 |
0.103070666 |
- |
- |
hypothetical protein CICLE_v10024045mg, partial [Citrus clementina] |
| 8 |
Hb_000197_260 |
0.1032517717 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |
| 9 |
Hb_000928_200 |
0.104420698 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 10 |
Hb_080601_010 |
0.1062527805 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 11 |
Hb_000646_040 |
0.1094858124 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74-like [Jatropha curcas] |
| 12 |
Hb_029791_010 |
0.111679715 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 13 |
Hb_002186_170 |
0.113948151 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 14 |
Hb_000599_160 |
0.1151166198 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 15 |
Hb_000098_230 |
0.1164683187 |
- |
- |
Potassium channel AKT2/3, putative [Ricinus communis] |
| 16 |
Hb_001541_170 |
0.11721705 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 17 |
Hb_152914_010 |
0.1184197932 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 18 |
Hb_007545_220 |
0.11846576 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 19 |
Hb_005183_070 |
0.1188396561 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 20 |
Hb_103225_030 |
0.1192311588 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |