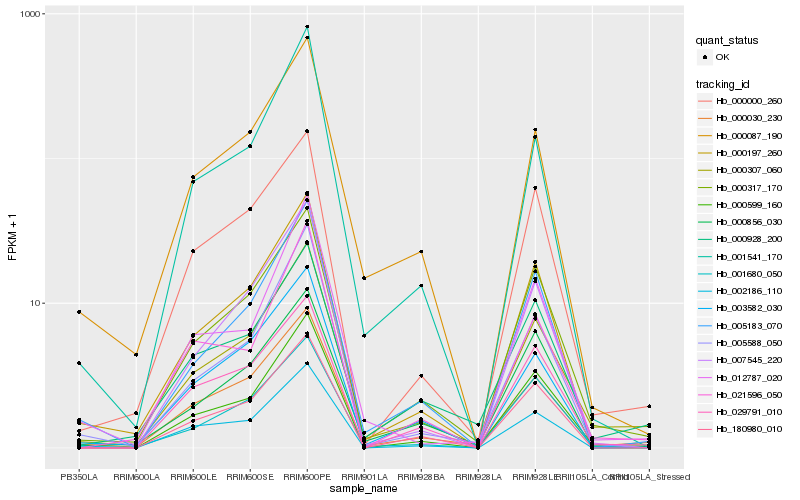
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000307_060 |
0.0 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 2 |
Hb_000030_230 |
0.0726259447 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 3 |
Hb_000317_170 |
0.0763565207 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 4 |
Hb_000599_160 |
0.0868864765 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 5 |
Hb_000928_200 |
0.1066842498 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 6 |
Hb_005183_070 |
0.1091344273 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 7 |
Hb_000000_260 |
0.1103687897 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 8 |
Hb_000197_260 |
0.1109676237 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |
| 9 |
Hb_000087_190 |
0.1119955489 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 10 |
Hb_001541_170 |
0.1185309789 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 11 |
Hb_003582_030 |
0.1194033992 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 12 |
Hb_007545_220 |
0.119585144 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 13 |
Hb_001680_050 |
0.1196282364 |
- |
- |
- |
| 14 |
Hb_029791_010 |
0.1208164927 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 15 |
Hb_005588_050 |
0.120963051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_021596_050 |
0.125981795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000856_030 |
0.1274791276 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_002186_110 |
0.1281314498 |
- |
- |
hypothetical protein CICLE_v10024045mg, partial [Citrus clementina] |
| 19 |
Hb_012787_020 |
0.1290523337 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 20 |
Hb_180980_010 |
0.1291991647 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |