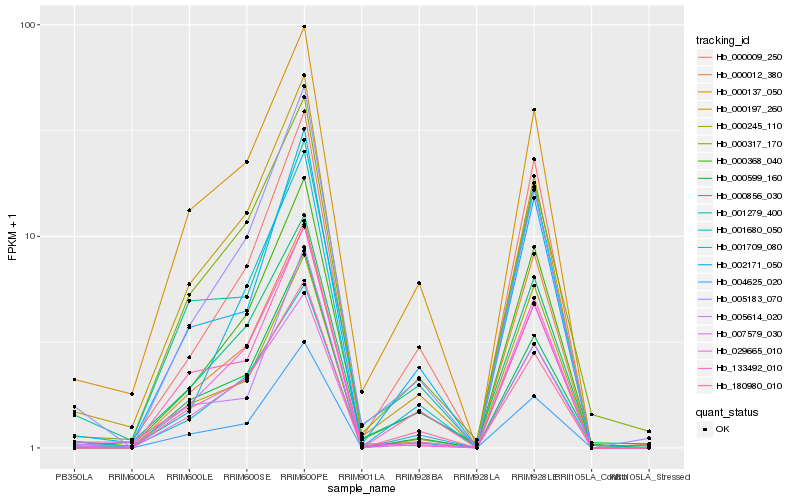
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000368_040 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000856_030 |
0.064364666 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_001680_050 |
0.0692543218 |
- |
- |
- |
| 4 |
Hb_005183_070 |
0.0848041362 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 5 |
Hb_000009_250 |
0.0876904818 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 6 |
Hb_029665_010 |
0.0884029025 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000317_170 |
0.0888951938 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 8 |
Hb_007579_030 |
0.0933375382 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 9 |
Hb_002171_050 |
0.0937925177 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 10 |
Hb_000197_260 |
0.0971589925 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |
| 11 |
Hb_180980_010 |
0.1073363038 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 12 |
Hb_001709_080 |
0.1084681505 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000137_050 |
0.1110744736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004625_020 |
0.1118867547 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 15 |
Hb_005614_020 |
0.1139386448 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000599_160 |
0.1156614168 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 17 |
Hb_000245_110 |
0.1157111028 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 18 |
Hb_000012_380 |
0.1173855709 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_133492_010 |
0.1189237052 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_001279_400 |
0.1197614256 |
- |
- |
PREDICTED: uncharacterized protein LOC105633544 [Jatropha curcas] |