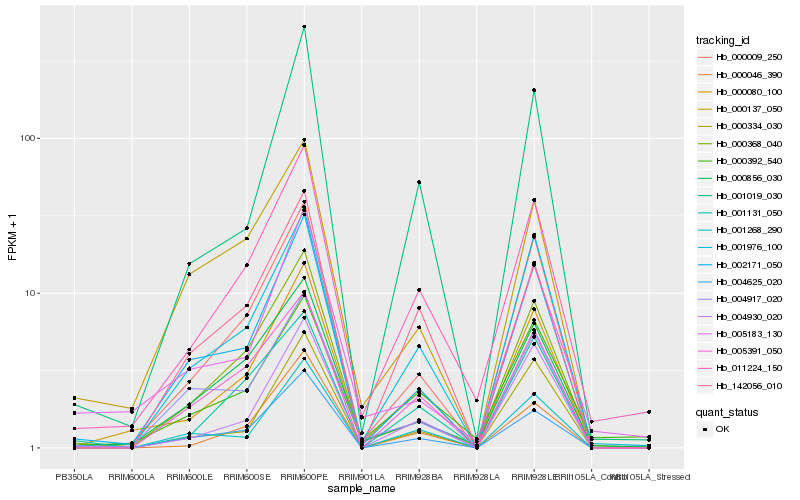
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000080_100 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 2 |
Hb_000009_250 |
0.0981137713 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 3 |
Hb_011224_150 |
0.1008714551 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001976_100 |
0.1112211135 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002171_050 |
0.1158210021 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase PEPR1 [Populus euphratica] |
| 6 |
Hb_004930_020 |
0.1190439587 |
transcription factor |
TF Family: MYB |
PREDICTED: transcriptional activator Myb-like [Jatropha curcas] |
| 7 |
Hb_001019_030 |
0.1233662631 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase 1-like [Jatropha curcas] |
| 8 |
Hb_004625_020 |
0.1263183544 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 9 |
Hb_000368_040 |
0.1275171687 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000046_390 |
0.1315307623 |
- |
- |
PREDICTED: CASP-like protein 1D1 [Jatropha curcas] |
| 11 |
Hb_000856_030 |
0.1329425403 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_005391_050 |
0.1332065257 |
- |
- |
PREDICTED: uncharacterized protein LOC105643654 [Jatropha curcas] |
| 13 |
Hb_000334_030 |
0.1347572139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_142056_010 |
0.1363775737 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001268_290 |
0.136735213 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 16 |
Hb_001131_050 |
0.1372800367 |
- |
- |
hypothetical protein EUGRSUZ_C02968 [Eucalyptus grandis] |
| 17 |
Hb_000137_050 |
0.1466907785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000392_540 |
0.1481473071 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Sesamum indicum] |
| 19 |
Hb_005183_130 |
0.1488078874 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 20 |
Hb_004917_020 |
0.1502751453 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF2.2-like isoform X2 [Populus euphratica] |