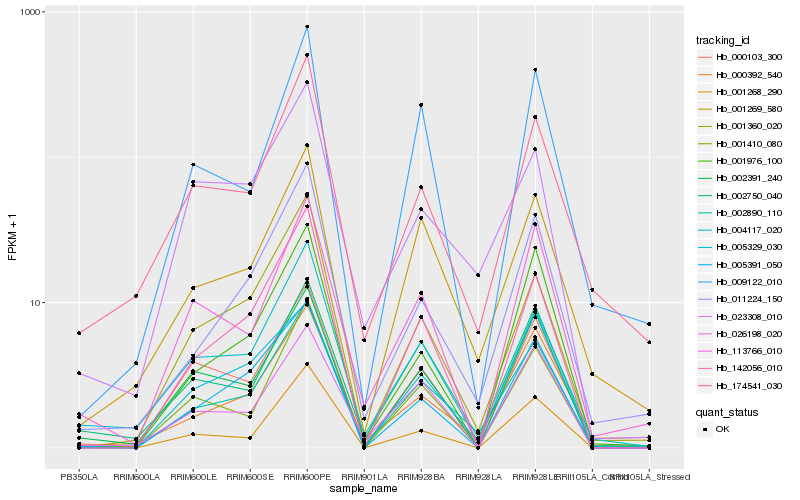
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_110 |
0.0 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 2 |
Hb_001269_580 |
0.102090859 |
- |
- |
PREDICTED: shikimate O-hydroxycinnamoyltransferase [Jatropha curcas] |
| 3 |
Hb_011224_150 |
0.1149262321 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000392_540 |
0.121852802 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Sesamum indicum] |
| 5 |
Hb_009122_010 |
0.1241339538 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_142056_010 |
0.125287938 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002750_040 |
0.1283280423 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 8 |
Hb_001360_020 |
0.1297614834 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 9 |
Hb_001410_080 |
0.129911054 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 10 |
Hb_000103_300 |
0.1338514676 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 11 |
Hb_001268_290 |
0.138660737 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 12 |
Hb_005391_050 |
0.1388084538 |
- |
- |
PREDICTED: uncharacterized protein LOC105643654 [Jatropha curcas] |
| 13 |
Hb_004117_020 |
0.1394292162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005329_030 |
0.1402128899 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 15 |
Hb_001976_100 |
0.1412803856 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_113766_010 |
0.141672981 |
- |
- |
plasma membrane H+ ATPase, partial [Nicotiana plumbaginifolia] |
| 17 |
Hb_174541_030 |
0.1418847833 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 18 |
Hb_002391_240 |
0.1434114976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_026198_020 |
0.1435614037 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 20 |
Hb_023308_010 |
0.1485057148 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |