| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
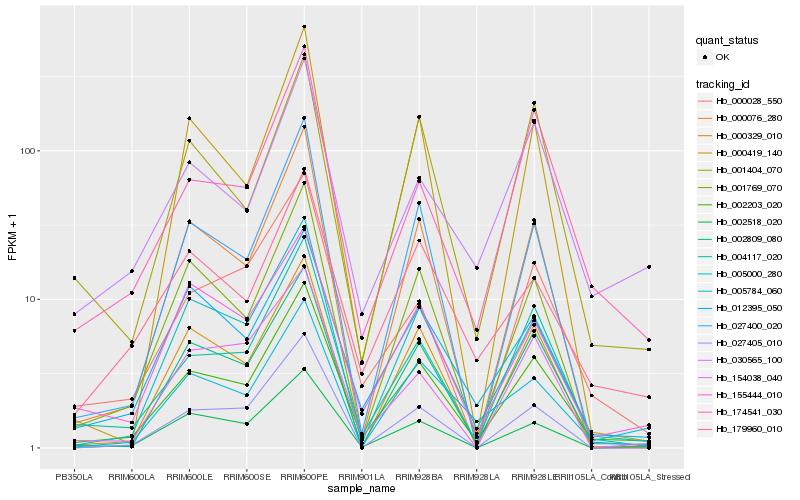
Hb_002203_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004117_020 |
0.0942876924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005000_280 |
0.0984721574 |
- |
- |
PREDICTED: uncharacterized protein LOC105637905 [Jatropha curcas] |
| 4 |
Hb_002809_080 |
0.100189736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000076_280 |
0.1126632813 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 6 |
Hb_001769_070 |
0.113182974 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_027400_020 |
0.115746754 |
- |
- |
PREDICTED: uncharacterized protein At5g22580 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002518_020 |
0.1184190772 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 9 |
Hb_000329_010 |
0.1188008233 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 10 |
Hb_012395_050 |
0.1209519879 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 11 |
Hb_174541_030 |
0.1238530803 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 12 |
Hb_000028_550 |
0.1249582283 |
- |
- |
unknown [Medicago truncatula] |
| 13 |
Hb_179960_010 |
0.129045124 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 14 |
Hb_155444_010 |
0.1294607266 |
- |
- |
- |
| 15 |
Hb_000419_140 |
0.1308432859 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 16 |
Hb_005784_060 |
0.1313255647 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 17 |
Hb_030565_100 |
0.1329903434 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 18 |
Hb_027405_010 |
0.1345354791 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 19 |
Hb_001404_070 |
0.1351161637 |
- |
- |
sucrose synthase 4 [Hevea brasiliensis] |
| 20 |
Hb_154038_040 |
0.1358949958 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |