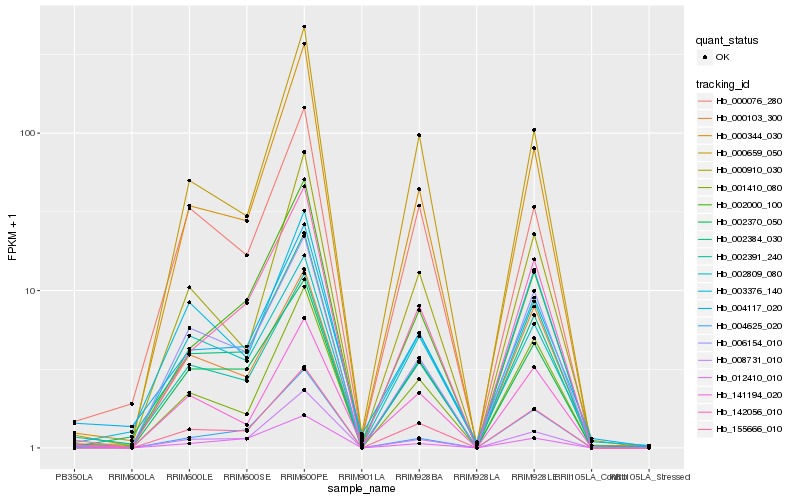
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_155666_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_01214 [Jatropha curcas] |
| 2 |
Hb_004117_020 |
0.1151875495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001410_080 |
0.1243574613 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 4 |
Hb_012410_010 |
0.1266291013 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 5 |
Hb_141194_020 |
0.1292569376 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 6 |
Hb_002391_240 |
0.1355479572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002370_050 |
0.1402812326 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 8 |
Hb_002384_030 |
0.140481345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000103_300 |
0.1410153243 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 10 |
Hb_006154_010 |
0.1423264165 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_004625_020 |
0.1424582102 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 12 |
Hb_142056_010 |
0.1440379241 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000076_280 |
0.1488466634 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 14 |
Hb_000659_050 |
0.1499327851 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 15 |
Hb_000910_030 |
0.1507371973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000344_030 |
0.1509960867 |
- |
- |
phenylalanine ammonia-lyase 1 [Manihot esculenta] |
| 17 |
Hb_002809_080 |
0.1532830017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003376_140 |
0.1547756381 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 19 |
Hb_002000_100 |
0.1562390868 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 20 |
Hb_008731_010 |
0.156880941 |
- |
- |
hypothetical protein POPTR_0605s00010g [Populus trichocarpa] |