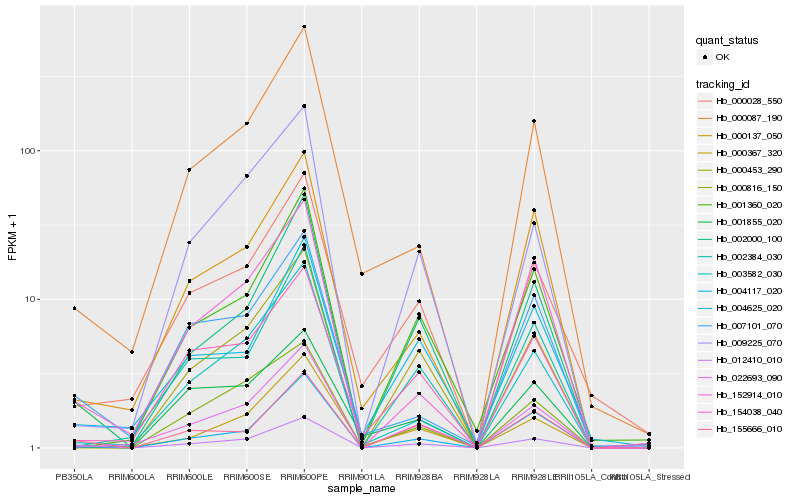
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012410_010 |
0.0 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_155666_010 |
0.1266291013 |
- |
- |
hypothetical protein JCGZ_01214 [Jatropha curcas] |
| 3 |
Hb_022693_090 |
0.1412807055 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_004625_020 |
0.1470569986 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4 [Jatropha curcas] |
| 5 |
Hb_152914_010 |
0.1496316603 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 6 |
Hb_000453_290 |
0.1527020446 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 7 |
Hb_000367_320 |
0.1555849908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003582_030 |
0.1587882446 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 9 |
Hb_000137_050 |
0.1595561101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_009225_070 |
0.1601104841 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 11 |
Hb_154038_040 |
0.1618832361 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 12 |
Hb_000087_190 |
0.1628798547 |
- |
- |
hypothetical protein CICLE_v10033278mg [Citrus clementina] |
| 13 |
Hb_007101_070 |
0.163583691 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 14 |
Hb_002000_100 |
0.1649930052 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 15 |
Hb_001360_020 |
0.1651894608 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |
| 16 |
Hb_002384_030 |
0.1655975274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000028_550 |
0.1657786273 |
- |
- |
unknown [Medicago truncatula] |
| 18 |
Hb_004117_020 |
0.1665503793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000816_150 |
0.167750484 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 20 |
Hb_001855_020 |
0.1700430573 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |