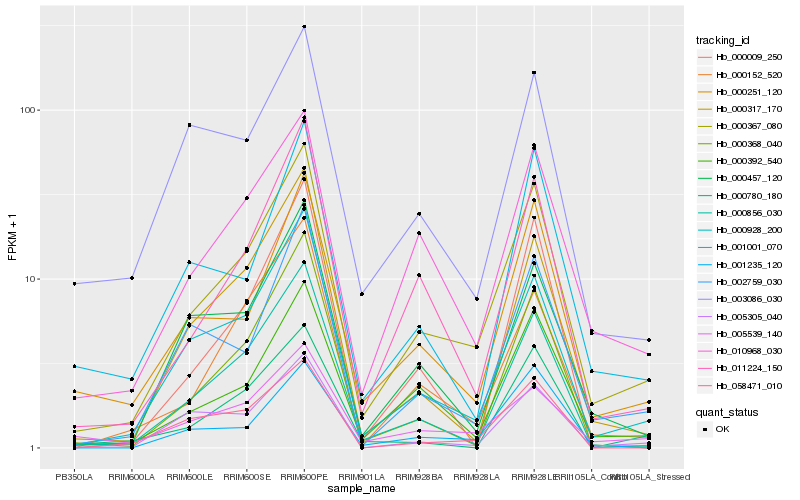
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_080 |
0.0 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 2 |
Hb_000928_200 |
0.1077496308 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 3 |
Hb_000392_540 |
0.1101460466 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Sesamum indicum] |
| 4 |
Hb_010968_030 |
0.1150247284 |
- |
- |
PREDICTED: uncharacterized protein LOC105643149 [Jatropha curcas] |
| 5 |
Hb_011224_150 |
0.1196423531 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At4g11680 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000856_030 |
0.1237701794 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_000251_120 |
0.1259008769 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 8 |
Hb_002759_030 |
0.1390930365 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 9 |
Hb_000457_120 |
0.1433908501 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 10 |
Hb_000152_520 |
0.1436240556 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 11 |
Hb_000317_170 |
0.1447844412 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 12 |
Hb_001001_070 |
0.1497284574 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 13 |
Hb_000368_040 |
0.1499468764 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_005305_040 |
0.1503549182 |
transcription factor |
TF Family: C3H |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_001235_120 |
0.1506165244 |
- |
- |
hypothetical protein JCGZ_03395 [Jatropha curcas] |
| 16 |
Hb_005539_140 |
0.1513597695 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 17 |
Hb_000009_250 |
0.1577796661 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 18 |
Hb_058471_010 |
0.1593194794 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 19 |
Hb_000780_180 |
0.1601826807 |
- |
- |
PREDICTED: uncharacterized protein LOC105641547 [Jatropha curcas] |
| 20 |
Hb_003086_030 |
0.1607196174 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |