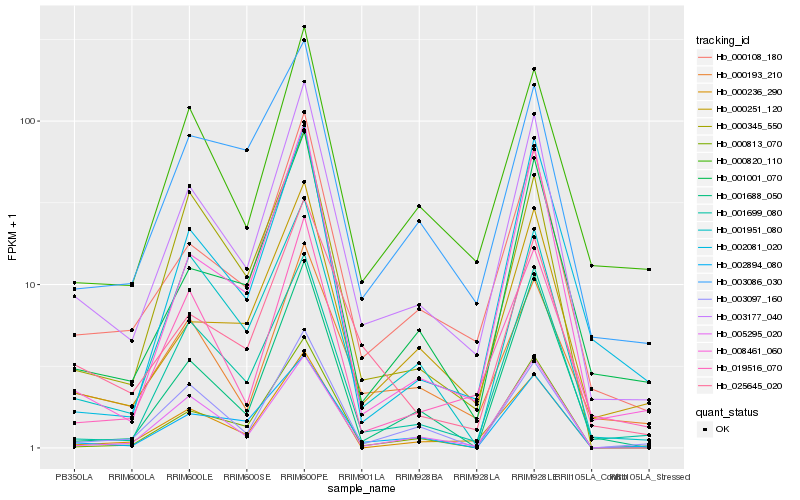
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003177_040 |
0.0 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 2 |
Hb_000108_180 |
0.0766245568 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 3 |
Hb_001001_070 |
0.1100559792 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 4 |
Hb_000236_290 |
0.1122243008 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 5 |
Hb_008461_060 |
0.1137452336 |
- |
- |
hypothetical protein JCGZ_17379 [Jatropha curcas] |
| 6 |
Hb_001699_080 |
0.1158348336 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_000251_120 |
0.1159009147 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 8 |
Hb_019516_070 |
0.1176298801 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 9 |
Hb_025645_020 |
0.1183661526 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 10 |
Hb_002894_080 |
0.1198147423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000820_110 |
0.1205309859 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 12 |
Hb_001951_080 |
0.1271031977 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000193_210 |
0.1292055705 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |
| 14 |
Hb_003097_160 |
0.1297215982 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein sel-10 [Jatropha curcas] |
| 15 |
Hb_000813_070 |
0.1334702822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001688_050 |
0.1347706208 |
- |
- |
PREDICTED: reticulon-like protein B9 [Jatropha curcas] |
| 17 |
Hb_000345_550 |
0.1351793169 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 18 |
Hb_003086_030 |
0.1357516925 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 19 |
Hb_005295_020 |
0.137718904 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 20 |
Hb_002081_020 |
0.1416085468 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |