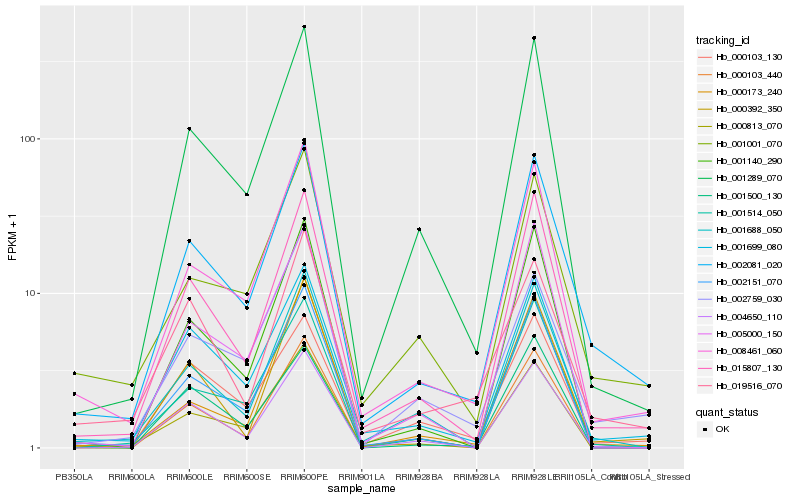
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002081_020 |
0.0 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 2 |
Hb_015807_130 |
0.0868889669 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008461_060 |
0.09179935 |
- |
- |
hypothetical protein JCGZ_17379 [Jatropha curcas] |
| 4 |
Hb_001699_080 |
0.0980434668 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_001289_070 |
0.1105597879 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 6 |
Hb_001001_070 |
0.1133384674 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 7 |
Hb_001688_050 |
0.1165698957 |
- |
- |
PREDICTED: reticulon-like protein B9 [Jatropha curcas] |
| 8 |
Hb_002151_070 |
0.1246113137 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 2-like [Jatropha curcas] |
| 9 |
Hb_000103_130 |
0.1255763895 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 10 |
Hb_004650_110 |
0.1268266812 |
- |
- |
hypothetical protein POPTR_0002s08760g [Populus trichocarpa] |
| 11 |
Hb_001514_050 |
0.127424575 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 12 |
Hb_001140_290 |
0.1327933283 |
- |
- |
hypothetical protein JCGZ_26107 [Jatropha curcas] |
| 13 |
Hb_000392_350 |
0.13338169 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_002759_030 |
0.1341661081 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |
| 15 |
Hb_019516_070 |
0.1341903665 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 16 |
Hb_005000_150 |
0.1367592048 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000813_070 |
0.1377927982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000173_240 |
0.1382684853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001500_130 |
0.1385520837 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 20 |
Hb_000103_440 |
0.1389296964 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |