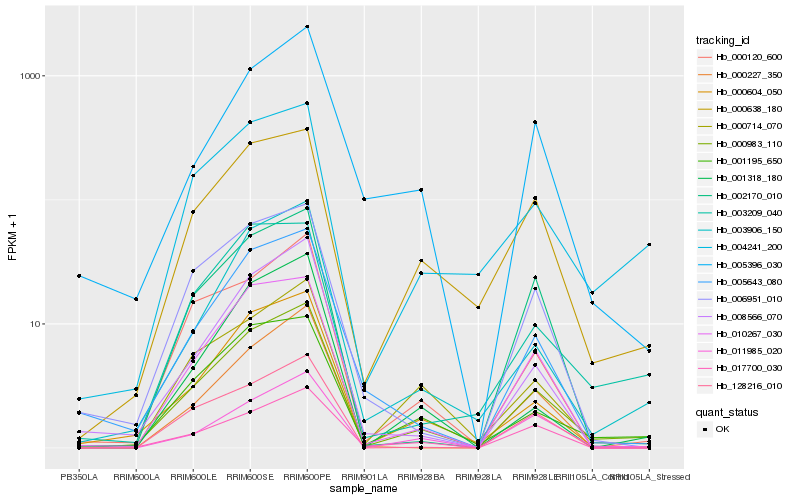
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000983_110 |
0.0 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 2 |
Hb_000604_050 |
0.1184442796 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 3 |
Hb_001195_650 |
0.1201700246 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 4 |
Hb_003906_150 |
0.120592867 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 5 |
Hb_010267_030 |
0.126772852 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000714_070 |
0.1330149509 |
transcription factor |
TF Family: LOB |
hypothetical protein JCGZ_14754 [Jatropha curcas] |
| 7 |
Hb_000120_600 |
0.1346097398 |
- |
- |
PREDICTED: uncharacterized protein LOC105628582 [Jatropha curcas] |
| 8 |
Hb_011985_020 |
0.1349314168 |
- |
- |
PREDICTED: phospholipase A1-Igamma3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005643_080 |
0.136245593 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 10 |
Hb_000638_180 |
0.1365013323 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 11 |
Hb_017700_030 |
0.138924594 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 12 |
Hb_008566_070 |
0.1397389208 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004241_200 |
0.1415354915 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 14 |
Hb_128216_010 |
0.142810532 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 15 |
Hb_002170_010 |
0.1435155723 |
- |
- |
hypothetical protein JCGZ_25046 [Jatropha curcas] |
| 16 |
Hb_003209_040 |
0.1457647494 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Gossypium raimondii] |
| 17 |
Hb_006951_010 |
0.1459853099 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 10-like [Populus euphratica] |
| 18 |
Hb_001318_180 |
0.1474467499 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 1 [Jatropha curcas] |
| 19 |
Hb_005396_030 |
0.1483296894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000227_350 |
0.1485068345 |
- |
- |
hypothetical protein CICLE_v10006537mg [Citrus clementina] |