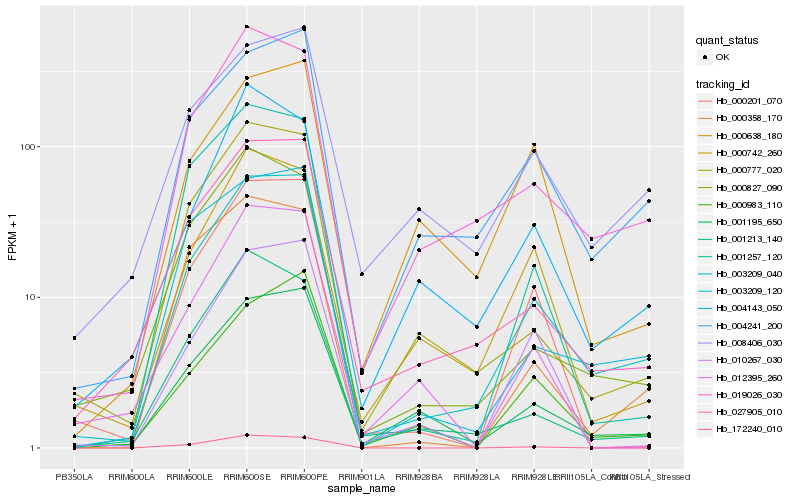
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003209_040 |
0.0 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Gossypium raimondii] |
| 2 |
Hb_027905_010 |
0.112068005 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 3 |
Hb_004241_200 |
0.1124764084 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 4 |
Hb_001195_650 |
0.1144070714 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 5 |
Hb_019026_030 |
0.117088023 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 6 |
Hb_003209_120 |
0.1246284555 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 7 |
Hb_008406_030 |
0.1352916311 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 8 |
Hb_000777_020 |
0.1380291972 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 9 |
Hb_000201_070 |
0.1401707183 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 10 |
Hb_000983_110 |
0.1457647494 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 11 |
Hb_000742_260 |
0.1457982132 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 12 |
Hb_000827_090 |
0.1501375081 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 13 |
Hb_001257_120 |
0.1549289518 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 14 |
Hb_000358_170 |
0.1556344183 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 15 |
Hb_001213_140 |
0.1564066303 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 16 |
Hb_004143_050 |
0.1571075513 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 17 |
Hb_010267_030 |
0.1599618334 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_172240_010 |
0.1617271107 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |
| 19 |
Hb_012395_260 |
0.1656182455 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA13 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000638_180 |
0.1666869687 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |