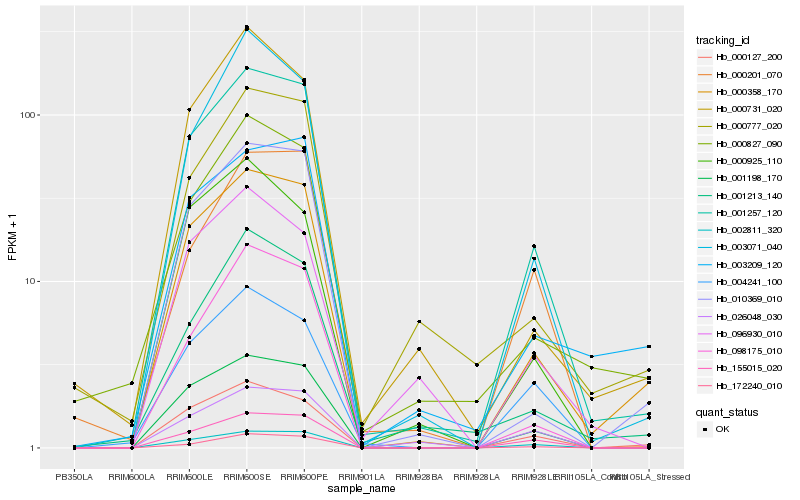
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001257_120 |
0.0 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 2 |
Hb_001198_170 |
0.06846729 |
- |
- |
- |
| 3 |
Hb_000358_170 |
0.0768547998 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 4 |
Hb_172240_010 |
0.0820578899 |
- |
- |
PREDICTED: uncharacterized protein LOC105795660 [Gossypium raimondii] |
| 5 |
Hb_002811_320 |
0.090234925 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 6 |
Hb_155015_020 |
0.0914616023 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 7 |
Hb_098175_010 |
0.1079549341 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 8 |
Hb_000127_200 |
0.110202073 |
- |
- |
PREDICTED: uncharacterized protein LOC100255537 [Vitis vinifera] |
| 9 |
Hb_010369_010 |
0.1144235348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003071_040 |
0.115340499 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF017 [Jatropha curcas] |
| 11 |
Hb_004241_100 |
0.1218303393 |
- |
- |
PREDICTED: transcription factor bHLH123-like isoform X1 [Populus euphratica] |
| 12 |
Hb_001213_140 |
0.1287581295 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 13 |
Hb_000827_090 |
0.1319784108 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 14 |
Hb_000777_020 |
0.1324502163 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 15 |
Hb_000201_070 |
0.1325312882 |
- |
- |
hypothetical protein POPTR_0067s00200g [Populus trichocarpa] |
| 16 |
Hb_000925_110 |
0.1377078202 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 17 |
Hb_000731_020 |
0.1385534239 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 18 |
Hb_026048_030 |
0.139327273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003209_120 |
0.1407502261 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 20 |
Hb_096930_010 |
0.1424402596 |
- |
- |
hypothetical protein JCGZ_27059 [Jatropha curcas] |