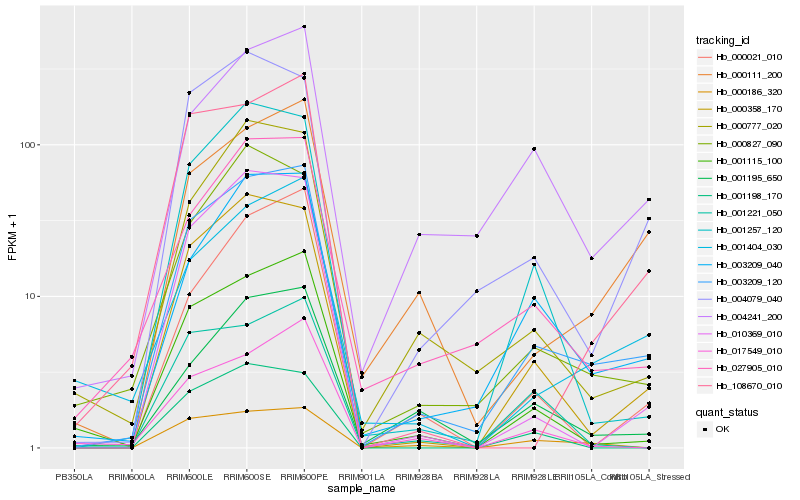
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003209_120 |
0.0 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 2 |
Hb_000358_170 |
0.1097174063 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 3 |
Hb_003209_040 |
0.1246284555 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Gossypium raimondii] |
| 4 |
Hb_001115_100 |
0.1259075808 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 5 |
Hb_027905_010 |
0.1353652496 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 6 |
Hb_000777_020 |
0.1389061591 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 7 |
Hb_001257_120 |
0.1407502261 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 9 [Jatropha curcas] |
| 8 |
Hb_010369_010 |
0.1412576127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001195_650 |
0.1440504905 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 10 |
Hb_001404_030 |
0.1441436702 |
- |
- |
DWNN domain isoform 3 [Theobroma cacao] |
| 11 |
Hb_108670_010 |
0.1464413815 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 12 |
Hb_004079_040 |
0.1464580445 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 13 |
Hb_000111_200 |
0.1481116615 |
- |
- |
PREDICTED: CYSTM1 family protein A-like [Cucumis melo] |
| 14 |
Hb_000827_090 |
0.1487612488 |
- |
- |
PREDICTED: uncharacterized protein LOC103714121 [Phoenix dactylifera] |
| 15 |
Hb_000021_010 |
0.1534780781 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000186_320 |
0.1587104131 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 17 |
Hb_004241_200 |
0.1628022819 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 18 |
Hb_017549_010 |
0.1645121415 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 19 |
Hb_001198_170 |
0.1680710308 |
- |
- |
- |
| 20 |
Hb_001221_050 |
0.168207348 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |