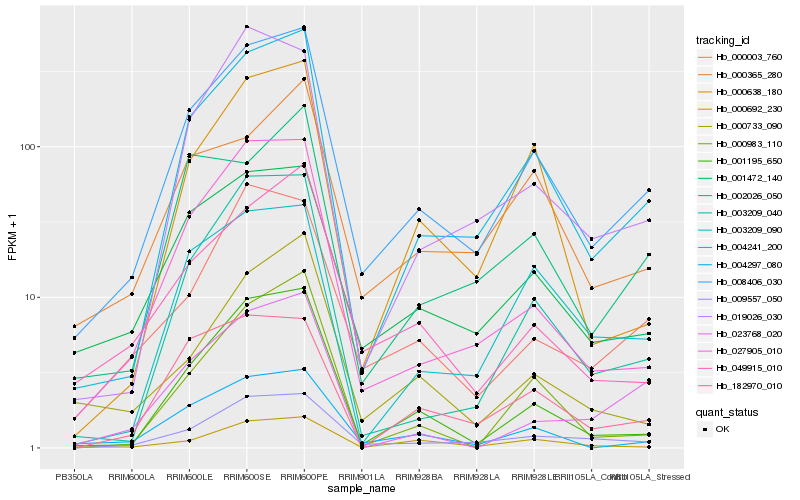
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008406_030 |
0.0 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 2 |
Hb_004241_200 |
0.073822092 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 3 |
Hb_009557_050 |
0.1114654957 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 4 |
Hb_001472_140 |
0.1209922751 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 5 |
Hb_001195_650 |
0.122346617 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 6 |
Hb_004297_080 |
0.1225246414 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 7 |
Hb_027905_010 |
0.1309330761 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 8 |
Hb_003209_040 |
0.1352916311 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Gossypium raimondii] |
| 9 |
Hb_049915_010 |
0.1423460225 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 10 |
Hb_000638_180 |
0.1443646924 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 11 |
Hb_000003_760 |
0.1483157145 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA26-like [Jatropha curcas] |
| 12 |
Hb_003209_090 |
0.1508324032 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase family protein [Theobroma cacao] |
| 13 |
Hb_019026_030 |
0.1550801629 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 14 |
Hb_000365_280 |
0.1557137868 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 15 |
Hb_182970_010 |
0.1563811762 |
- |
- |
calmodulin-binding protein 60-D [Populus trichocarpa] |
| 16 |
Hb_000692_230 |
0.1569601637 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 17 |
Hb_000983_110 |
0.1627768824 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 18 |
Hb_000733_090 |
0.1629909563 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 19 |
Hb_002026_050 |
0.1635108953 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 20 |
Hb_023768_020 |
0.1679839833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |