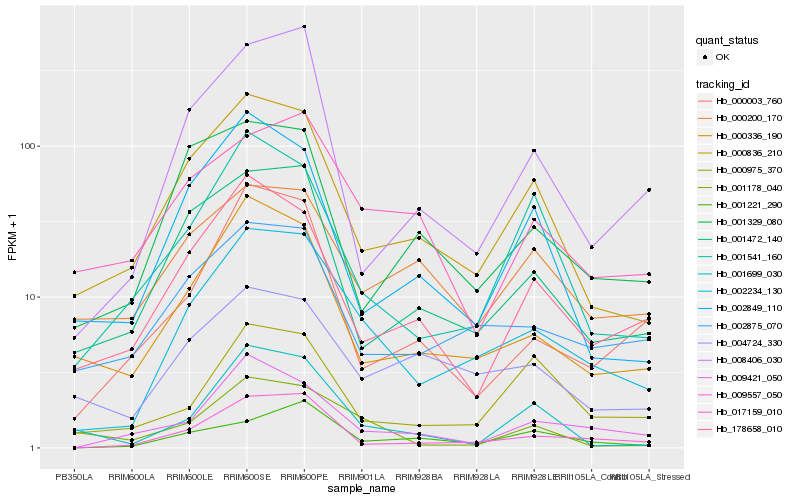
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002234_130 |
0.0 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 2 |
Hb_009557_050 |
0.1484870528 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 3 |
Hb_000836_210 |
0.1649958668 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 4 |
Hb_002875_070 |
0.1690657585 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 5 |
Hb_000336_190 |
0.1699631911 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 6 |
Hb_001472_140 |
0.1811208217 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 7 |
Hb_009421_050 |
0.1870637479 |
- |
- |
PREDICTED: uncharacterized protein LOC105647037 [Jatropha curcas] |
| 8 |
Hb_008406_030 |
0.1938348007 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 9 |
Hb_178658_010 |
0.1947792972 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 10 |
Hb_004724_330 |
0.195181314 |
- |
- |
Capsanthin/capsorubin synthase, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_001541_160 |
0.1962051762 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 12 |
Hb_001221_290 |
0.2010005814 |
- |
- |
hypothetical protein RCOM_1506010 [Ricinus communis] |
| 13 |
Hb_000003_760 |
0.2019989225 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA26-like [Jatropha curcas] |
| 14 |
Hb_017159_010 |
0.2031800033 |
- |
- |
- |
| 15 |
Hb_001178_040 |
0.2053287769 |
desease resistance |
Gene Name: ABC_trans_N |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 16 |
Hb_002849_110 |
0.2080194557 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 17 |
Hb_001699_030 |
0.208959302 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000975_370 |
0.209355142 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000200_170 |
0.2099711802 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 20 |
Hb_001329_080 |
0.2109769611 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |