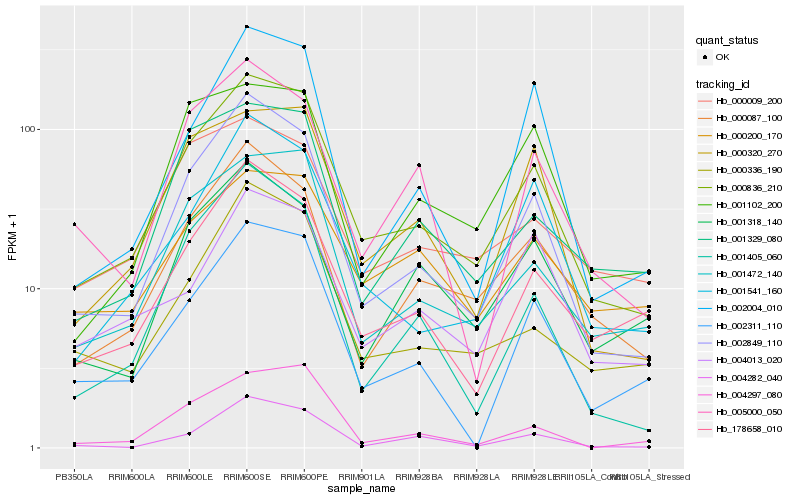
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_210 |
0.0 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 2 |
Hb_002849_110 |
0.0812270331 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 3 |
Hb_001472_140 |
0.1060329637 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 4 |
Hb_000087_100 |
0.1166239299 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 5 |
Hb_178658_010 |
0.1220426552 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 6 |
Hb_001541_160 |
0.1246047014 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 7 |
Hb_002311_110 |
0.1270478315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000336_190 |
0.1278271975 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 9 |
Hb_000320_270 |
0.1295116223 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001329_080 |
0.1357567453 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 11 |
Hb_001405_060 |
0.1440354015 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 12 |
Hb_002004_010 |
0.1452952538 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 13 |
Hb_001102_200 |
0.1466906264 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 14 |
Hb_001318_140 |
0.1474668806 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 15 |
Hb_005000_050 |
0.1485006106 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 16 |
Hb_000009_200 |
0.1490105541 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 17 |
Hb_000200_170 |
0.1501474057 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 18 |
Hb_004282_040 |
0.150499643 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_004297_080 |
0.1516236799 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 20 |
Hb_004013_020 |
0.1520738098 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |