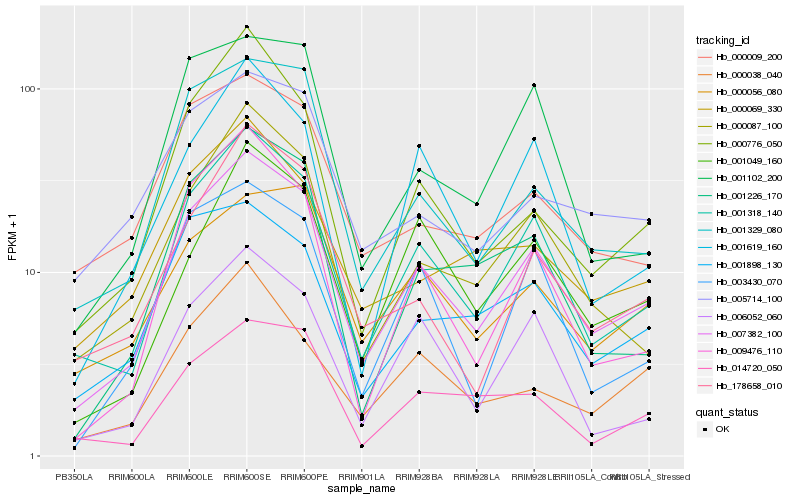
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007382_100 |
0.0 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 2 |
Hb_001318_140 |
0.0656723794 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 3 |
Hb_000087_100 |
0.11460445 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 4 |
Hb_001329_080 |
0.1195549638 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 5 |
Hb_001619_160 |
0.1255602453 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 6 |
Hb_001898_130 |
0.1265177262 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_003430_070 |
0.1277243736 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 8 |
Hb_001049_160 |
0.1298877143 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 9 |
Hb_001226_170 |
0.1299790691 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 10 |
Hb_178658_010 |
0.1300630647 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 11 |
Hb_009476_110 |
0.1304827888 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 12 |
Hb_000776_050 |
0.1317891949 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 13 |
Hb_006052_060 |
0.1325155864 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 14 |
Hb_000056_080 |
0.1331887539 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000069_330 |
0.1333847108 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_014720_050 |
0.1334460009 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 17 |
Hb_000038_040 |
0.1355813636 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 18 |
Hb_000009_200 |
0.1361900553 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 19 |
Hb_001102_200 |
0.1364044703 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 20 |
Hb_005714_100 |
0.1370656418 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |