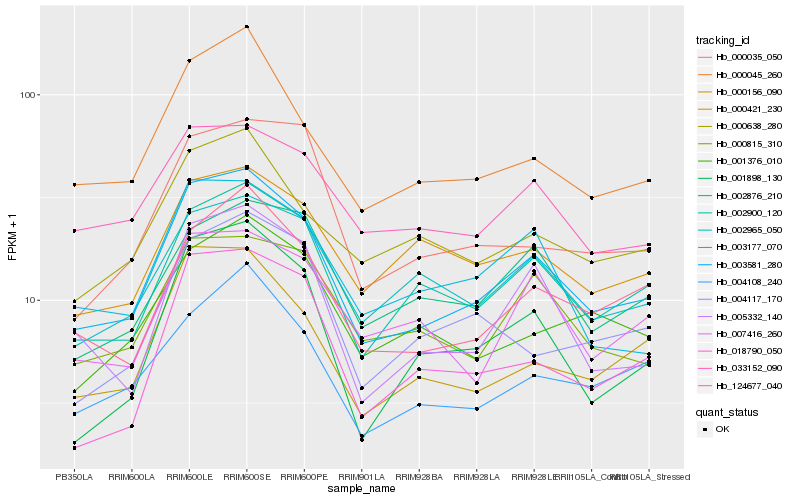
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003581_280 |
0.0 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 2 |
Hb_002900_120 |
0.084018519 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 3 |
Hb_000045_260 |
0.090263171 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_124677_040 |
0.0905796931 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 5 |
Hb_000421_230 |
0.0936755134 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 6 |
Hb_005332_140 |
0.098769875 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X2 [Jatropha curcas] |
| 7 |
Hb_004108_240 |
0.1039155141 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 8 |
Hb_000815_310 |
0.1067941476 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 9 |
Hb_002965_050 |
0.1074633424 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 10 |
Hb_003177_070 |
0.1077428376 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 11 |
Hb_033152_090 |
0.1079719753 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 12 |
Hb_000638_280 |
0.1088002621 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 13 |
Hb_000035_050 |
0.1109435228 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 14 |
Hb_001898_130 |
0.1110738671 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_018790_050 |
0.1121779553 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 16 |
Hb_000156_090 |
0.1151046753 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001376_010 |
0.1154166316 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 18 |
Hb_002876_210 |
0.1156678447 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 19 |
Hb_007416_260 |
0.1159148187 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF12-A-like [Jatropha curcas] |
| 20 |
Hb_004117_170 |
0.1173870536 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |