| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
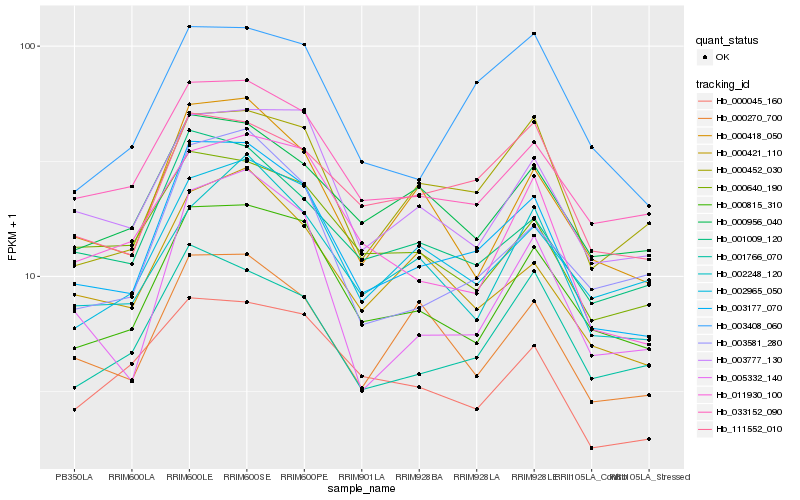
Hb_003177_070 |
0.0 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 2 |
Hb_000815_310 |
0.088757223 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 3 |
Hb_001009_120 |
0.0914792572 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 4 |
Hb_033152_090 |
0.0930135626 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 5 |
Hb_000418_050 |
0.0935367968 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 6 |
Hb_000421_110 |
0.0980789138 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_003777_130 |
0.0999611131 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 8 |
Hb_000270_700 |
0.1043383404 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 9 |
Hb_003581_280 |
0.1077428376 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 10 |
Hb_005332_140 |
0.1081376948 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000956_040 |
0.1082331643 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 12 |
Hb_111552_010 |
0.1087561359 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 13 |
Hb_011930_100 |
0.1088268143 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 14 |
Hb_001766_070 |
0.1089078288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000640_190 |
0.1104292197 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000452_030 |
0.1108539979 |
- |
- |
Protein phosphatase 1 regulatory subunit, putative [Ricinus communis] |
| 17 |
Hb_002965_050 |
0.1112837128 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 18 |
Hb_002248_120 |
0.1114719845 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 19 |
Hb_000045_160 |
0.1122112249 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 20 |
Hb_003408_060 |
0.113293366 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |