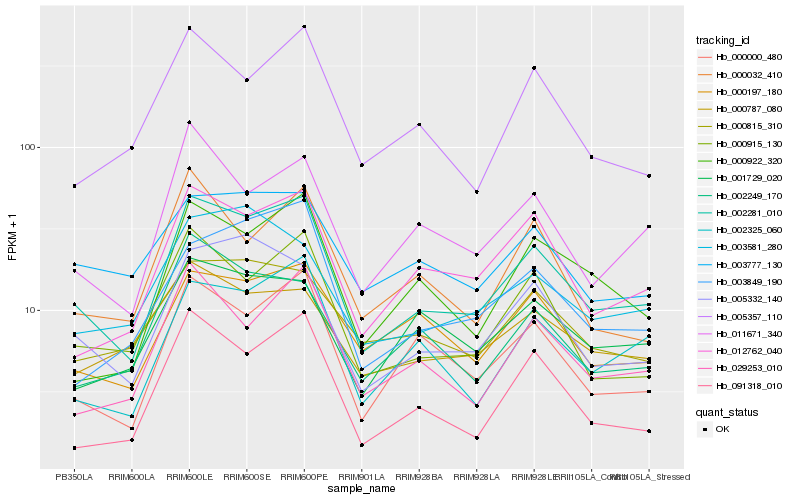
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002281_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s47550g [Populus trichocarpa] |
| 2 |
Hb_000915_130 |
0.1132613401 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 3 |
Hb_012762_040 |
0.1141710428 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 4 |
Hb_005332_140 |
0.1152787453 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000000_480 |
0.1156868523 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002325_060 |
0.1178216961 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 7 |
Hb_011671_340 |
0.1271481823 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 8 |
Hb_000922_320 |
0.1290264996 |
- |
- |
hypothetical protein B456_001G071400 [Gossypium raimondii] |
| 9 |
Hb_029253_010 |
0.1292798494 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 10 |
Hb_005357_110 |
0.1295015765 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 11 |
Hb_000197_180 |
0.1297578366 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 12 |
Hb_003777_130 |
0.1343824752 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 13 |
Hb_003849_190 |
0.1346394843 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 14 |
Hb_091318_010 |
0.1348197195 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 15 |
Hb_000815_310 |
0.1379039005 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000787_080 |
0.1390602834 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 17 |
Hb_000032_410 |
0.1401532626 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 18 |
Hb_002249_170 |
0.1407605547 |
- |
- |
copine, putative [Ricinus communis] |
| 19 |
Hb_003581_280 |
0.1409404736 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 20 |
Hb_001729_020 |
0.14159479 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |