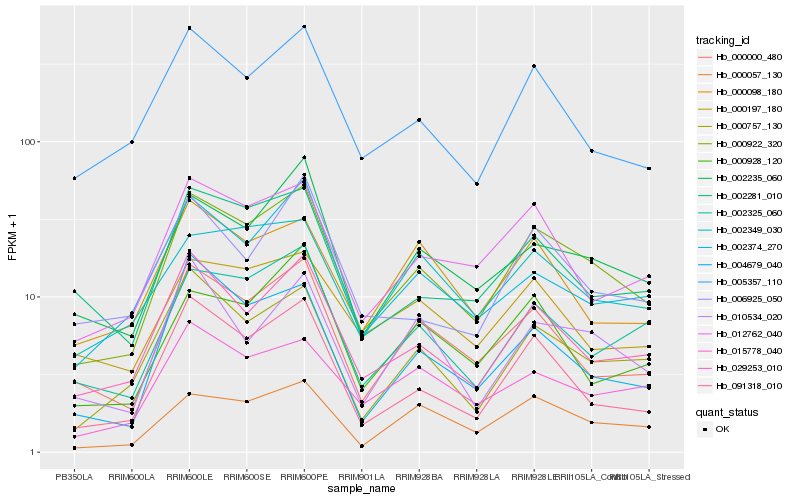
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_320 |
0.0 |
- |
- |
hypothetical protein B456_001G071400 [Gossypium raimondii] |
| 2 |
Hb_029253_010 |
0.1045746195 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 3 |
Hb_000000_480 |
0.1120088548 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002235_060 |
0.1132438178 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_002325_060 |
0.1152292714 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 6 |
Hb_000757_130 |
0.1155598447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_012762_040 |
0.1159753887 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 8 |
Hb_000057_130 |
0.11654983 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 9 |
Hb_005357_110 |
0.1168784661 |
- |
- |
hypothetical protein B456_002G083000 [Gossypium raimondii] |
| 10 |
Hb_091318_010 |
0.1172653975 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 11 |
Hb_002374_270 |
0.1284393019 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002281_010 |
0.1290264996 |
- |
- |
hypothetical protein POPTR_0001s47550g [Populus trichocarpa] |
| 13 |
Hb_015778_040 |
0.1307036701 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 14 |
Hb_000197_180 |
0.1358478101 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 15 |
Hb_000928_120 |
0.1383587622 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 16 |
Hb_002349_030 |
0.1384951493 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004679_040 |
0.1385890638 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 18 |
Hb_000098_180 |
0.1415921965 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 19 |
Hb_010534_020 |
0.141680501 |
- |
- |
PREDICTED: protein XRI1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006925_050 |
0.1419259515 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |