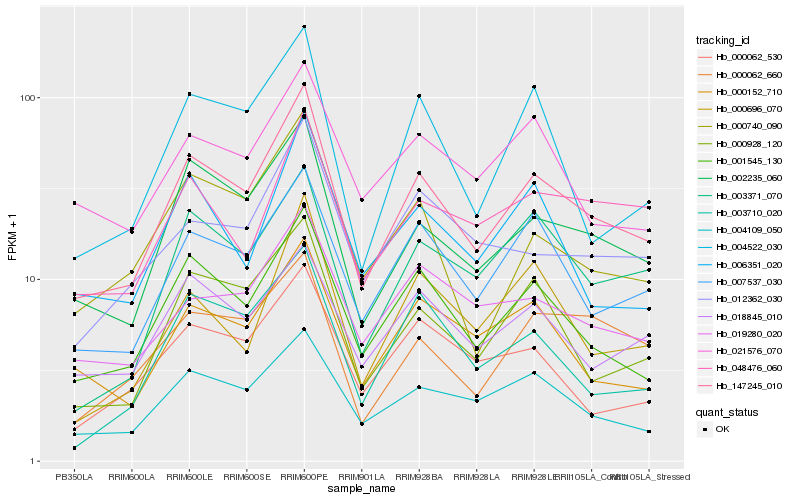
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_147245_010 |
0.0 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 2 |
Hb_002235_060 |
0.085328979 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 3 |
Hb_001545_130 |
0.0867830161 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 4 |
Hb_007537_030 |
0.1026141209 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000928_120 |
0.1054379734 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 6 |
Hb_004109_050 |
0.1062424047 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 7 |
Hb_000152_710 |
0.1200291419 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 1 [Jatropha curcas] |
| 8 |
Hb_003371_070 |
0.1204622474 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000740_090 |
0.1213210066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_018845_010 |
0.1218309892 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 11 |
Hb_004522_030 |
0.123893918 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 12 |
Hb_019280_020 |
0.1240093732 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012362_030 |
0.1287661782 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 14 |
Hb_003710_020 |
0.1296529003 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 15 |
Hb_048476_060 |
0.1305498432 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000062_660 |
0.1306543448 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 17 |
Hb_006351_020 |
0.1309285628 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 18 |
Hb_000062_530 |
0.1327271849 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 19 |
Hb_021576_070 |
0.1330612584 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 20 |
Hb_000696_070 |
0.1333353594 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |