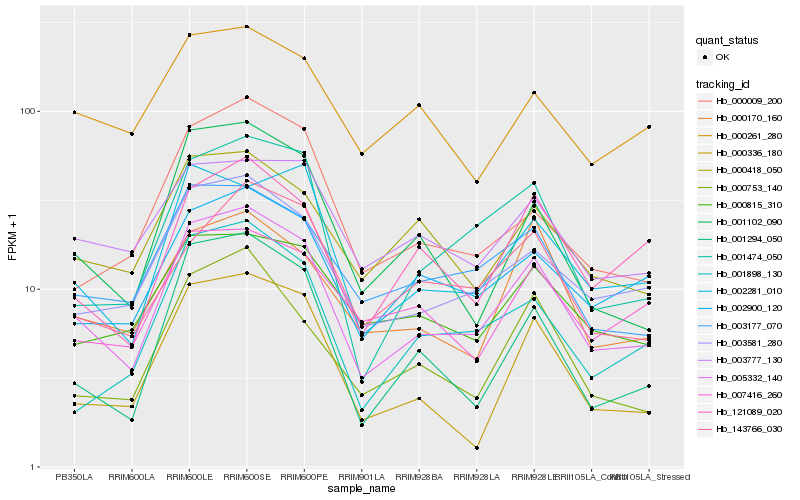
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005332_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X2 [Jatropha curcas] |
| 2 |
Hb_003581_280 |
0.098769875 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 3 |
Hb_003177_070 |
0.1081376948 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 4 |
Hb_001102_090 |
0.1133906229 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 5 |
Hb_001474_050 |
0.1150255392 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 6 |
Hb_002281_010 |
0.1152787453 |
- |
- |
hypothetical protein POPTR_0001s47550g [Populus trichocarpa] |
| 7 |
Hb_002900_120 |
0.1194389802 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 8 |
Hb_000753_140 |
0.1196587395 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 9 |
Hb_000418_050 |
0.1222254511 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 10 |
Hb_001294_050 |
0.1229451041 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 11 |
Hb_000170_160 |
0.1266427948 |
- |
- |
PREDICTED: NAD kinase 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_121089_020 |
0.1273915147 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 13 |
Hb_000336_180 |
0.1275616418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000815_310 |
0.1276161779 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 15 |
Hb_001898_130 |
0.1282751084 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_003777_130 |
0.1286108978 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |
| 17 |
Hb_143766_030 |
0.1292477494 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000009_200 |
0.1310323128 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 19 |
Hb_007416_260 |
0.1374026496 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF12-A-like [Jatropha curcas] |
| 20 |
Hb_000261_280 |
0.1382979133 |
- |
- |
BnaA10g23810D [Brassica napus] |