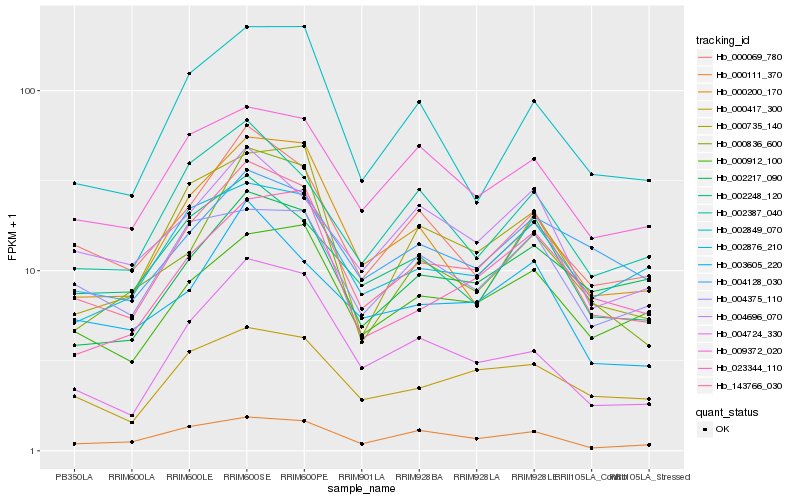
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143766_030 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_002248_120 |
0.1031379837 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 3 |
Hb_003605_220 |
0.1055809466 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 4 |
Hb_000200_170 |
0.1089589334 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 5 |
Hb_000735_140 |
0.1099767472 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 6 |
Hb_023344_110 |
0.1115125611 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 7 |
Hb_000069_780 |
0.1115377749 |
- |
- |
atpob1, putative [Ricinus communis] |
| 8 |
Hb_002876_210 |
0.1134396685 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 9 |
Hb_000912_100 |
0.1135207369 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 10 |
Hb_000111_370 |
0.1148363076 |
- |
- |
PREDICTED: cation/H(+) antiporter 2-like [Jatropha curcas] |
| 11 |
Hb_002217_090 |
0.1177425951 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 12 |
Hb_002849_070 |
0.118143555 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000417_300 |
0.1187282208 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 [Jatropha curcas] |
| 14 |
Hb_002387_040 |
0.1190299179 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 15 |
Hb_004375_110 |
0.1195472645 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX37 [Jatropha curcas] |
| 16 |
Hb_004696_070 |
0.1195597724 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 17 |
Hb_004128_030 |
0.1206439549 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 8-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_009372_020 |
0.1208348375 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 19 |
Hb_000836_600 |
0.1213220729 |
- |
- |
PREDICTED: delta(8)-fatty-acid desaturase 2 [Jatropha curcas] |
| 20 |
Hb_004724_330 |
0.1215236087 |
- |
- |
Capsanthin/capsorubin synthase, chloroplast precursor, putative [Ricinus communis] |