| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
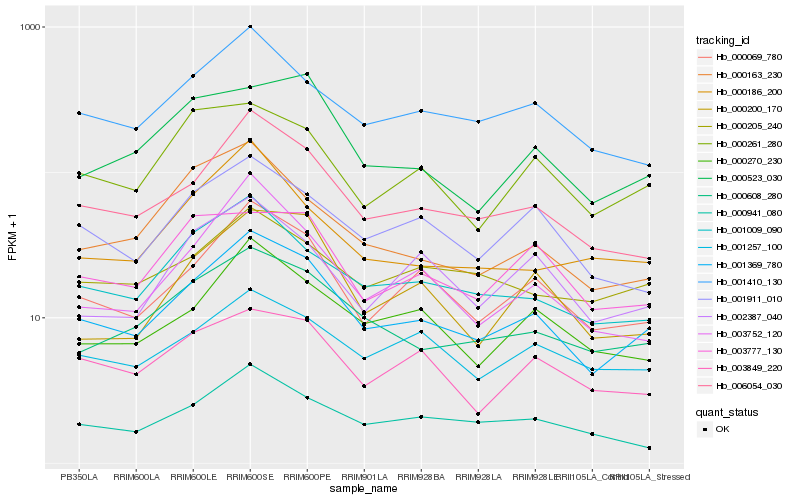
Hb_001369_780 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006054_030 |
0.0801937499 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 3 |
Hb_000069_780 |
0.0833250134 |
- |
- |
atpob1, putative [Ricinus communis] |
| 4 |
Hb_000608_280 |
0.0982279018 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 5 |
Hb_001009_090 |
0.1027880886 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 6 |
Hb_001410_130 |
0.1065341895 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 7 |
Hb_000270_230 |
0.109778475 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 8 |
Hb_000941_080 |
0.1169567271 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X3 [Populus euphratica] |
| 9 |
Hb_001911_010 |
0.1197141351 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 10 |
Hb_001257_100 |
0.1213510682 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 11 |
Hb_000261_280 |
0.1220609537 |
- |
- |
BnaA10g23810D [Brassica napus] |
| 12 |
Hb_003849_220 |
0.1231259279 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 13 |
Hb_003752_120 |
0.1235989086 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000200_170 |
0.1257238627 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 15 |
Hb_000523_030 |
0.1262915543 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 16 |
Hb_000163_230 |
0.1280058285 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 17 |
Hb_000186_200 |
0.128088957 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 18 |
Hb_000205_240 |
0.1281459154 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_002387_040 |
0.1281616512 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 20 |
Hb_003777_130 |
0.1292441043 |
- |
- |
PREDICTED: uncharacterized protein LOC105640939 [Jatropha curcas] |