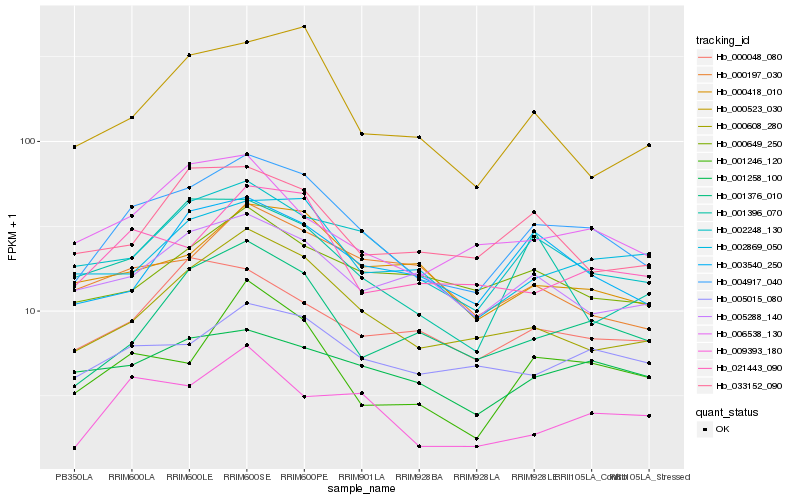
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004917_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 2 |
Hb_002248_130 |
0.1072215272 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 3 |
Hb_000608_280 |
0.109174216 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 4 |
Hb_003540_250 |
0.1245109494 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 5 |
Hb_021443_090 |
0.1258608802 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 6 |
Hb_005015_080 |
0.1275228357 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001376_010 |
0.129423637 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 8 |
Hb_000418_010 |
0.1299199274 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 9 |
Hb_005288_140 |
0.131692279 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 10 |
Hb_002869_050 |
0.1324473611 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 11 |
Hb_001246_120 |
0.1342322082 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |
| 12 |
Hb_001258_100 |
0.1360616203 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_033152_090 |
0.1368448403 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 14 |
Hb_009393_180 |
0.137358886 |
- |
- |
PREDICTED: uncharacterized protein LOC101292333 [Fragaria vesca subsp. vesca] |
| 15 |
Hb_000649_250 |
0.1387265742 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 16 |
Hb_006538_130 |
0.1388638237 |
- |
- |
Uncharacterized protein TCM_009867 [Theobroma cacao] |
| 17 |
Hb_000523_030 |
0.1399778402 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 18 |
Hb_000048_080 |
0.1400774641 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001396_070 |
0.1426505837 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000197_030 |
0.142872124 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |