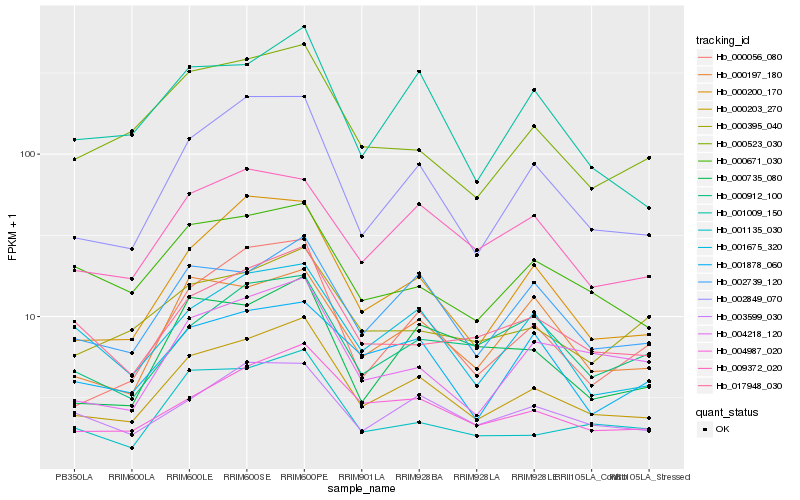
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000203_270 |
0.0 |
- |
- |
alpha-xylosidase, putative [Ricinus communis] |
| 2 |
Hb_004987_020 |
0.0819886744 |
- |
- |
- |
| 3 |
Hb_002849_070 |
0.0899025218 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002739_120 |
0.094042461 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 5 |
Hb_000200_170 |
0.100693488 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 6 |
Hb_001135_030 |
0.1031077052 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 7 |
Hb_003599_030 |
0.1058147019 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 8 |
Hb_000523_030 |
0.1058880069 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 9 |
Hb_001009_150 |
0.1059132596 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase [Gossypium arboreum] |
| 10 |
Hb_017948_030 |
0.1061533866 |
- |
- |
PREDICTED: uncharacterized protein LOC105639493 [Jatropha curcas] |
| 11 |
Hb_009372_020 |
0.107826551 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 12 |
Hb_004218_120 |
0.1094505944 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000197_180 |
0.1100924593 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 14 |
Hb_001675_320 |
0.1101405099 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 15 |
Hb_000056_080 |
0.1109356122 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000671_030 |
0.1119524296 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 17 |
Hb_000395_040 |
0.1143964105 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 18 |
Hb_000912_100 |
0.1173520697 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 19 |
Hb_001878_060 |
0.1175747366 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0007s15110g [Populus trichocarpa] |
| 20 |
Hb_000735_080 |
0.1178888943 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |