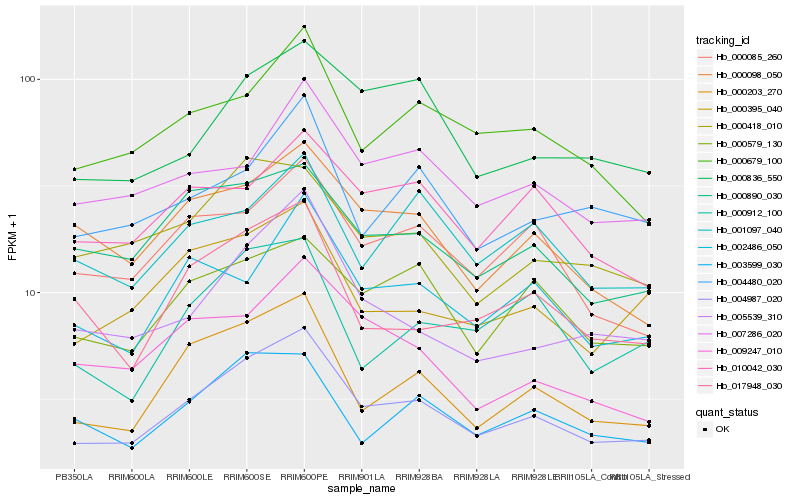
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004987_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000203_270 |
0.0819886744 |
- |
- |
alpha-xylosidase, putative [Ricinus communis] |
| 3 |
Hb_005539_310 |
0.1048498373 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000679_100 |
0.1071126863 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000085_260 |
0.1074217594 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 6 |
Hb_000395_040 |
0.1075587447 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 7 |
Hb_000890_030 |
0.1102729009 |
- |
- |
plastid CUL1 [Hevea brasiliensis] |
| 8 |
Hb_002486_050 |
0.1109341833 |
- |
- |
Multiple inositol polyphosphate phosphatase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_003599_030 |
0.1110598303 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000836_550 |
0.1118241334 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 11 |
Hb_017948_030 |
0.1122938261 |
- |
- |
PREDICTED: uncharacterized protein LOC105639493 [Jatropha curcas] |
| 12 |
Hb_000098_050 |
0.1125996945 |
- |
- |
BnaCnng11900D [Brassica napus] |
| 13 |
Hb_000579_130 |
0.1128162987 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_000418_010 |
0.1139256713 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 15 |
Hb_007286_020 |
0.1178679675 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 16 |
Hb_009247_010 |
0.1192917843 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004480_020 |
0.1193582972 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 18 |
Hb_000912_100 |
0.1203081455 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 19 |
Hb_001097_040 |
0.1213255876 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 20 |
Hb_010042_030 |
0.1226547303 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |