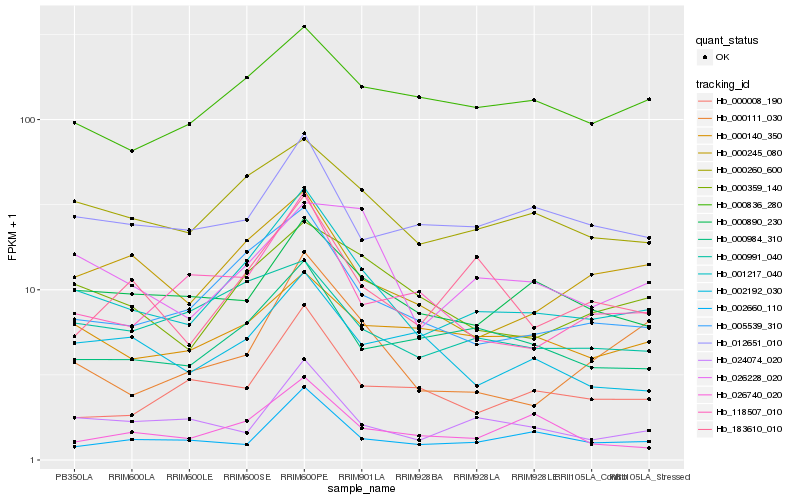
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001217_040 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 2 |
Hb_005539_310 |
0.0978992677 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000260_600 |
0.1174245225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000359_140 |
0.137295703 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 5 |
Hb_000984_310 |
0.1375767949 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 6 |
Hb_000245_080 |
0.1424934819 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-15 [Jatropha curcas] |
| 7 |
Hb_000890_230 |
0.1483938999 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 8 |
Hb_000991_040 |
0.150830264 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 9 |
Hb_024074_020 |
0.150959658 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 10 |
Hb_183610_010 |
0.1509793066 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 11 |
Hb_000836_280 |
0.1524000617 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_118507_010 |
0.1542182969 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 13 |
Hb_012651_010 |
0.1564249061 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 14 |
Hb_000140_350 |
0.1574815557 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_026740_020 |
0.1583342883 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 16 |
Hb_000008_190 |
0.1584087482 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002660_110 |
0.1584964554 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_026228_020 |
0.1585882191 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 19 |
Hb_000111_030 |
0.1588278694 |
- |
- |
PREDICTED: uncharacterized protein LOC104094030 [Nicotiana tomentosiformis] |
| 20 |
Hb_002192_030 |
0.1597717899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |