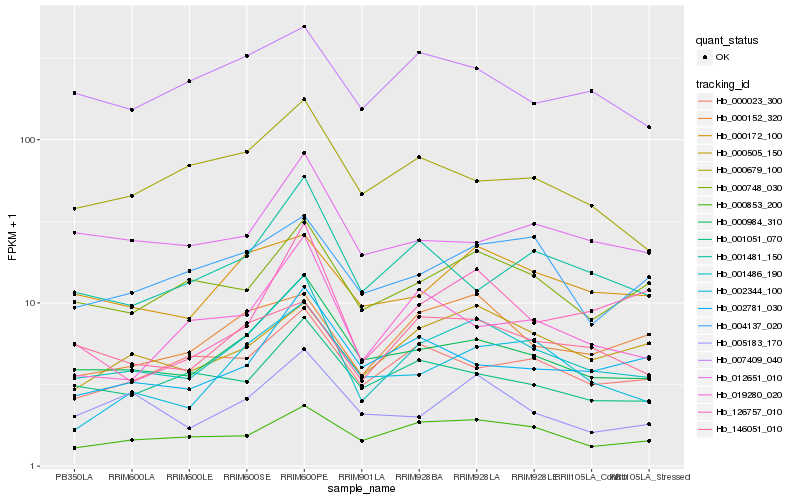
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001486_190 |
0.0 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 2 |
Hb_000984_310 |
0.0962954188 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 3 |
Hb_000748_030 |
0.1262094648 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_000853_200 |
0.1303463453 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 5 |
Hb_005183_170 |
0.1325182313 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g04860 [Jatropha curcas] |
| 6 |
Hb_146051_010 |
0.1345934523 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 7 |
Hb_000172_100 |
0.1351955424 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 2, peroxisomal [Jatropha curcas] |
| 8 |
Hb_000152_320 |
0.1383807846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000505_150 |
0.1393020008 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22-like [Jatropha curcas] |
| 10 |
Hb_002344_100 |
0.1394003615 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 11 |
Hb_001051_070 |
0.1418212878 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_019280_020 |
0.1422114233 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001481_150 |
0.1441145021 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 14 |
Hb_000023_300 |
0.1441847043 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 15 |
Hb_012651_010 |
0.144447867 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 16 |
Hb_000679_100 |
0.1449511206 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 17 |
Hb_126757_010 |
0.1471250059 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 18 |
Hb_004137_020 |
0.1485155635 |
- |
- |
PREDICTED: putative UDP-glucuronate:xylan alpha-glucuronosyltransferase 3 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007409_040 |
0.1489553916 |
- |
- |
actin family protein [Populus trichocarpa] |
| 20 |
Hb_002781_030 |
0.1496592489 |
- |
- |
protein binding protein, putative [Ricinus communis] |