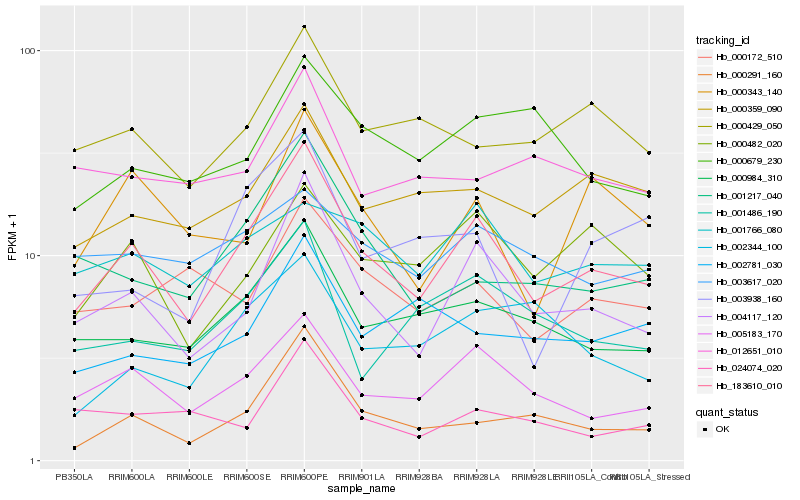
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183610_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 2 |
Hb_004117_120 |
0.094728476 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 3 |
Hb_005183_170 |
0.1137266251 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g04860 [Jatropha curcas] |
| 4 |
Hb_000984_310 |
0.1285217718 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 5 |
Hb_001217_040 |
0.1509793066 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 6 |
Hb_000291_160 |
0.1514817188 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 7 |
Hb_000482_020 |
0.1522002489 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 8 |
Hb_000359_090 |
0.1531251644 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 9 |
Hb_000429_050 |
0.1564933764 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000343_140 |
0.1578365472 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 11 |
Hb_001486_190 |
0.1618425565 |
- |
- |
hypothetical protein JCGZ_02702 [Jatropha curcas] |
| 12 |
Hb_000172_510 |
0.1682007114 |
- |
- |
hypothetical protein B456_007G243100 [Gossypium raimondii] |
| 13 |
Hb_001766_080 |
0.1711239666 |
- |
- |
PREDICTED: uncharacterized protein LOC105644125 [Jatropha curcas] |
| 14 |
Hb_000679_230 |
0.1718350937 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 15 |
Hb_003617_020 |
0.1720585883 |
- |
- |
PREDICTED: uncharacterized protein LOC105638179 [Jatropha curcas] |
| 16 |
Hb_003938_160 |
0.174698174 |
- |
- |
PREDICTED: transcription factor bHLH155 [Jatropha curcas] |
| 17 |
Hb_002781_030 |
0.1763288164 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_012651_010 |
0.1763569263 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 19 |
Hb_002344_100 |
0.1776123231 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 20 |
Hb_024074_020 |
0.1793238157 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |