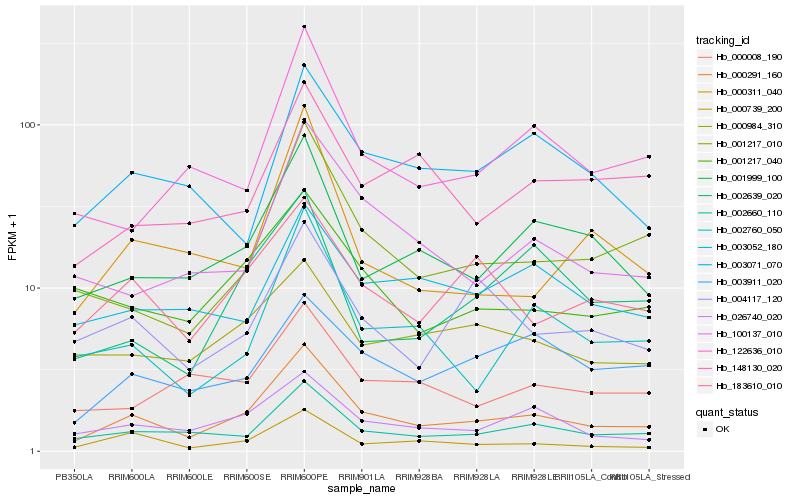
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000291_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 2 |
Hb_002660_110 |
0.133015186 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000739_200 |
0.1393907228 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 4 |
Hb_026740_020 |
0.144408207 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 5 |
Hb_001999_100 |
0.1472421689 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 6 |
Hb_001217_010 |
0.1484508649 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |
| 7 |
Hb_183610_010 |
0.1514817188 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 8 |
Hb_002760_050 |
0.1541033316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_100137_010 |
0.1599047128 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 10 |
Hb_001217_040 |
0.1610399762 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 11 |
Hb_004117_120 |
0.165237976 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 12 |
Hb_122636_010 |
0.1657172785 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 13 |
Hb_003911_020 |
0.167928122 |
- |
- |
PREDICTED: uncharacterized protein LOC105121332 [Populus euphratica] |
| 14 |
Hb_000984_310 |
0.1725599463 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 15 |
Hb_000008_190 |
0.1726045284 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_148130_020 |
0.1767143 |
- |
- |
- |
| 17 |
Hb_003052_180 |
0.1773989344 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 18 |
Hb_003071_070 |
0.1780848833 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |
| 19 |
Hb_002639_020 |
0.1796178112 |
- |
- |
PREDICTED: probable alpha-amylase 2 [Jatropha curcas] |
| 20 |
Hb_000311_040 |
0.1816084189 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |