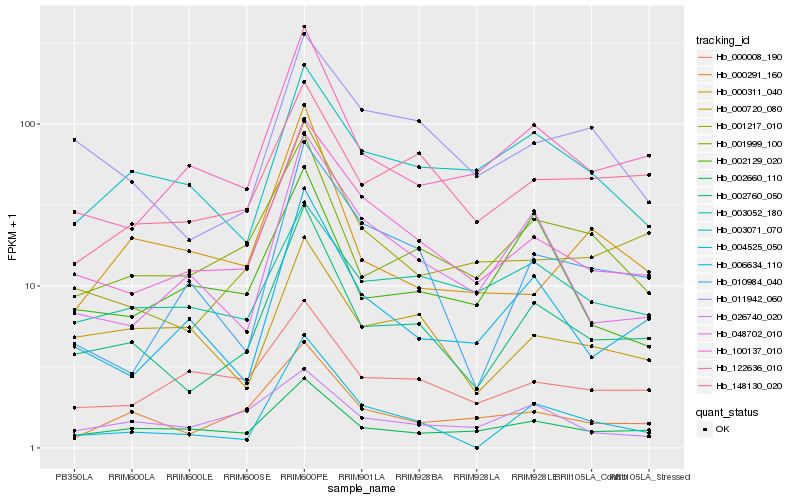
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002760_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_100137_010 |
0.1394953825 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 3 |
Hb_001217_010 |
0.1441420537 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |
| 4 |
Hb_001999_100 |
0.1447639395 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 5 |
Hb_122636_010 |
0.1484888053 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 6 |
Hb_000291_160 |
0.1541033316 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 7 |
Hb_002660_110 |
0.1555049397 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_148130_020 |
0.163618166 |
- |
- |
- |
| 9 |
Hb_004525_050 |
0.1637803164 |
- |
- |
similar to Drosophila melanogaster CG10960, partial [Drosophila yakuba] |
| 10 |
Hb_006634_110 |
0.1680770369 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 11 |
Hb_011942_060 |
0.1705286402 |
- |
- |
hypothetical protein CISIN_1g028965mg [Citrus sinensis] |
| 12 |
Hb_048702_010 |
0.1838190302 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_000720_080 |
0.1843272238 |
- |
- |
PREDICTED: bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 [Jatropha curcas] |
| 14 |
Hb_003071_070 |
0.1901897859 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |
| 15 |
Hb_000008_190 |
0.1902067131 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002129_020 |
0.1909111225 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 17 |
Hb_003052_180 |
0.1913819579 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 18 |
Hb_010984_040 |
0.1947319887 |
- |
- |
hypothetical protein M569_17286, partial [Genlisea aurea] |
| 19 |
Hb_000311_040 |
0.1949063219 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 20 |
Hb_026740_020 |
0.1959543955 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |