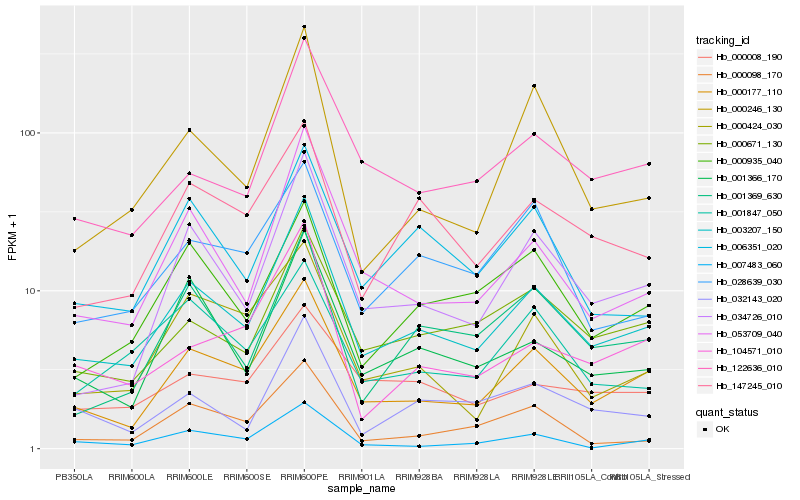
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003207_150 |
0.0 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 2 |
Hb_000177_110 |
0.0812404747 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 3 |
Hb_053709_040 |
0.1056169173 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 4 |
Hb_034726_010 |
0.1145018738 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X2 [Jatropha curcas] |
| 5 |
Hb_001366_170 |
0.1259422669 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 6 |
Hb_000671_130 |
0.1260185057 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_122636_010 |
0.1266982728 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 8 |
Hb_147245_010 |
0.1344632215 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 9 |
Hb_032143_020 |
0.1367424248 |
- |
- |
hypothetical protein POPTR_0009s06310g [Populus trichocarpa] |
| 10 |
Hb_006351_020 |
0.1392067829 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 11 |
Hb_001369_630 |
0.1397942479 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 12 |
Hb_000246_130 |
0.1426651929 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 13 |
Hb_104571_010 |
0.1471174511 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |
| 14 |
Hb_000935_040 |
0.1476442133 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 15 |
Hb_007483_060 |
0.1484856689 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 16 |
Hb_000424_030 |
0.1489729167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000008_190 |
0.1501038259 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000098_170 |
0.1501176028 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 19 |
Hb_001847_050 |
0.1530231159 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 20 |
Hb_028639_030 |
0.1533727808 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |