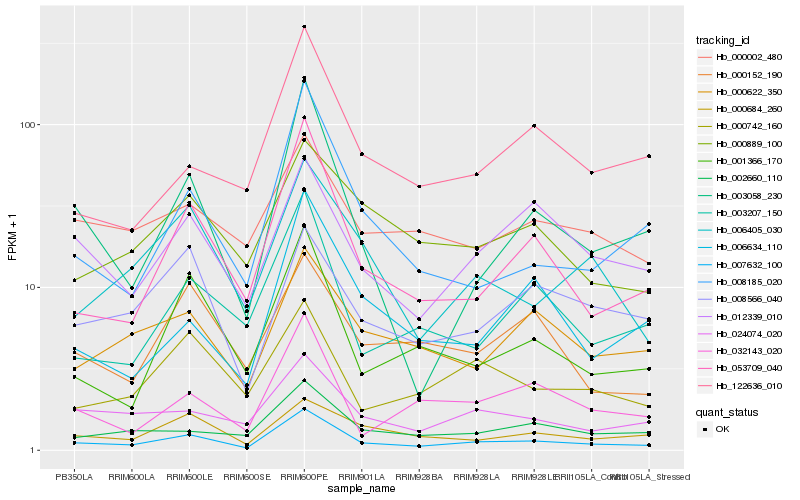
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007632_100 |
0.0 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 2 |
Hb_053709_040 |
0.1328524292 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 3 |
Hb_024074_020 |
0.1545010211 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 4 |
Hb_003207_150 |
0.1557949832 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 5 |
Hb_001366_170 |
0.156295605 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 6 |
Hb_003058_230 |
0.1578097859 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 7 |
Hb_002660_110 |
0.1580852663 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_012339_010 |
0.1585976354 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_032143_020 |
0.1596131453 |
- |
- |
hypothetical protein POPTR_0009s06310g [Populus trichocarpa] |
| 10 |
Hb_008185_020 |
0.1599047446 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 11 |
Hb_000622_350 |
0.1607047222 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 12 |
Hb_122636_010 |
0.1628645579 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 13 |
Hb_000684_260 |
0.1653344251 |
- |
- |
PREDICTED: probable DNA helicase MCM8 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000742_160 |
0.1656079916 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g20940 [Jatropha curcas] |
| 15 |
Hb_006634_110 |
0.1674834413 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 16 |
Hb_006405_030 |
0.1676234544 |
- |
- |
hypothetical protein VITISV_043238 [Vitis vinifera] |
| 17 |
Hb_008566_040 |
0.1716948836 |
- |
- |
PREDICTED: uncharacterized protein LOC105647590 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000889_100 |
0.1740627714 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 19 |
Hb_000002_480 |
0.1753504425 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000152_190 |
0.1764320012 |
- |
- |
PREDICTED: K(+) efflux antiporter 5 [Jatropha curcas] |