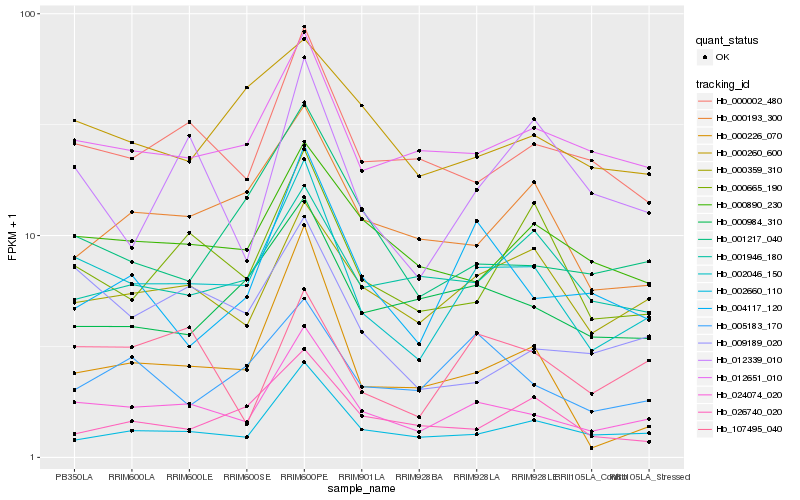
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002046_150 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g15080 [Jatropha curcas] |
| 2 |
Hb_024074_020 |
0.0797942127 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 3 |
Hb_012651_010 |
0.1409163168 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 4 |
Hb_000665_190 |
0.1442546741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000359_310 |
0.1451246287 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 6 |
Hb_000002_480 |
0.1491416167 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000193_300 |
0.1521635882 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |
| 8 |
Hb_002660_110 |
0.1535779673 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000890_230 |
0.1574190362 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 10 |
Hb_001217_040 |
0.1601481516 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 11 |
Hb_000984_310 |
0.1604739027 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 12 |
Hb_005183_170 |
0.1647304361 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g04860 [Jatropha curcas] |
| 13 |
Hb_009189_020 |
0.1675406223 |
- |
- |
PREDICTED: CASP-like protein 4B4 [Jatropha curcas] |
| 14 |
Hb_012339_010 |
0.1692026616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000260_600 |
0.1697359054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001946_180 |
0.170108715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004117_120 |
0.1706799075 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 18 |
Hb_026740_020 |
0.175840829 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 19 |
Hb_000226_070 |
0.1763144581 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 20 |
Hb_107495_040 |
0.1772325184 |
- |
- |
PREDICTED: 12-oxophytodienoate reductase 2-like [Citrus sinensis] |