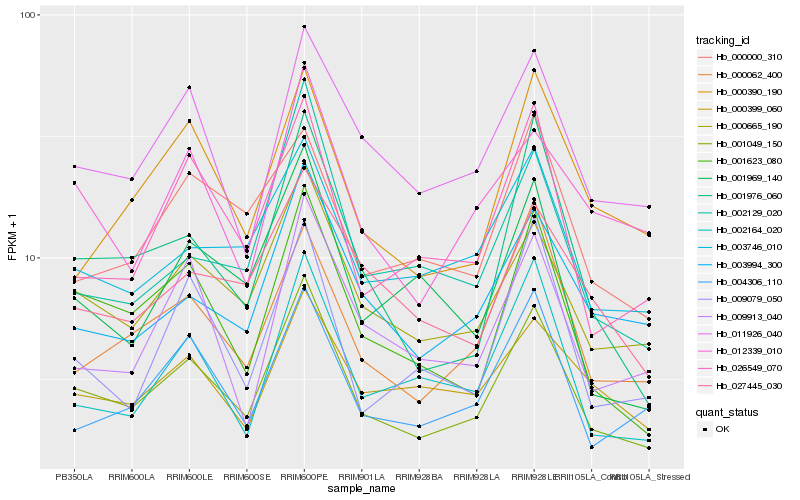
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001049_150 |
0.0 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 2 |
Hb_000665_190 |
0.0805803649 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001623_080 |
0.0875525738 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_011926_040 |
0.1095974063 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 5 |
Hb_026549_070 |
0.1125436707 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 6 |
Hb_000062_400 |
0.1151216205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001976_060 |
0.1157611602 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003994_300 |
0.1164927687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003746_010 |
0.117276928 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004306_110 |
0.1181706047 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 11 |
Hb_002164_020 |
0.1223046777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001969_140 |
0.1247036037 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 13 |
Hb_012339_010 |
0.1262793566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_027445_030 |
0.1270627119 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_002129_020 |
0.1285824829 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 16 |
Hb_009079_050 |
0.1291706106 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 17 |
Hb_009913_040 |
0.1297351152 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 18 |
Hb_000399_060 |
0.1303682948 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 19 |
Hb_000390_190 |
0.1304035262 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_000000_310 |
0.1308093193 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |