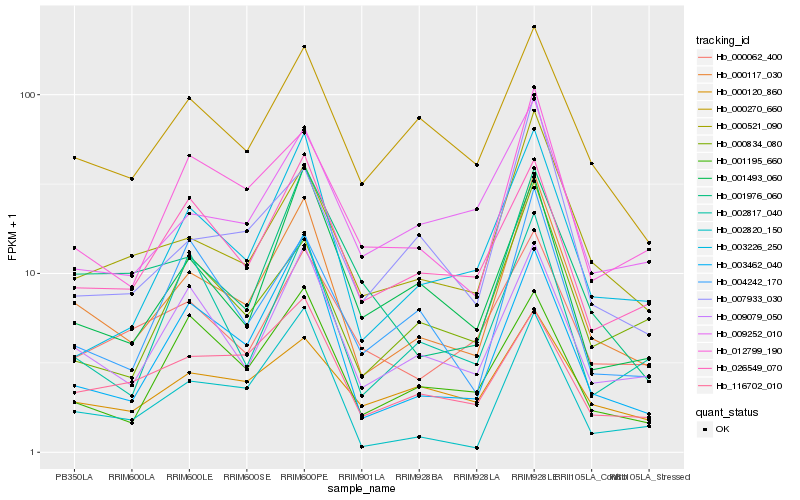
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000117_030 |
0.0 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 2 |
Hb_000521_090 |
0.1137114338 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009079_050 |
0.1182100267 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 4 |
Hb_003462_040 |
0.1322901277 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_012799_190 |
0.134194022 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 6 |
Hb_003226_250 |
0.139510671 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 7 |
Hb_000270_660 |
0.1417634565 |
- |
- |
purine permease, putative [Ricinus communis] |
| 8 |
Hb_001493_060 |
0.1435017729 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 9 |
Hb_002820_150 |
0.149258629 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_007933_030 |
0.1494853298 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 11 |
Hb_026549_070 |
0.149864537 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 12 |
Hb_001976_060 |
0.1508272846 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 13 |
Hb_009252_010 |
0.1521356105 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 14 |
Hb_000120_860 |
0.1524899196 |
- |
- |
nucellin, putative [Ricinus communis] |
| 15 |
Hb_000834_080 |
0.1531652557 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 16 |
Hb_004242_170 |
0.1539615103 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 17 |
Hb_000062_400 |
0.154190189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_116702_010 |
0.1542389468 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 19 |
Hb_001195_660 |
0.1562345178 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 20 |
Hb_002817_040 |
0.1564570386 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |