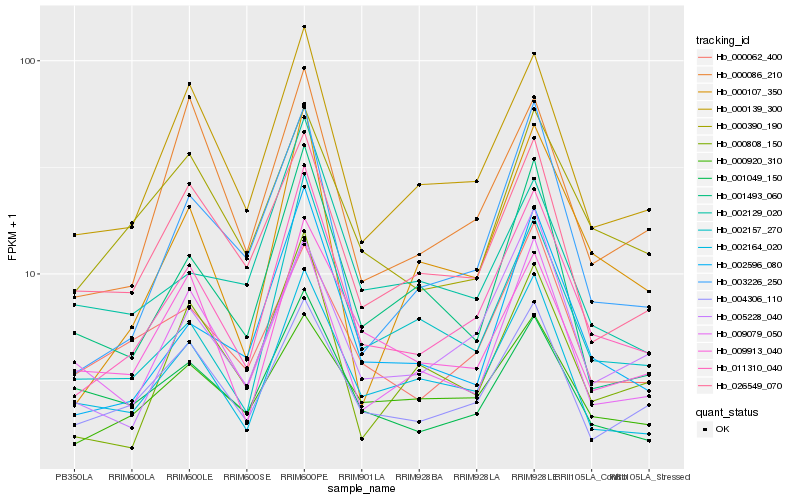
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011310_040 |
0.0 |
transcription factor |
TF Family: bZIP |
transcription factor hy5, putative [Ricinus communis] |
| 2 |
Hb_002596_080 |
0.0987278178 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 3 |
Hb_000139_300 |
0.1024084093 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 4 |
Hb_002157_270 |
0.1117354711 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 5 |
Hb_003226_250 |
0.1140597024 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 6 |
Hb_002164_020 |
0.1142175692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004306_110 |
0.1220692937 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 8 |
Hb_000107_350 |
0.1249119931 |
- |
- |
PREDICTED: uncharacterized protein LOC105635576 [Jatropha curcas] |
| 9 |
Hb_005228_040 |
0.1286619884 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 10 |
Hb_001493_060 |
0.1300637062 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 11 |
Hb_000808_150 |
0.1327827769 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 12 |
Hb_026549_070 |
0.1340510335 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 13 |
Hb_000390_190 |
0.1346931718 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000920_310 |
0.1354196152 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 15 |
Hb_009913_040 |
0.1365138896 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 16 |
Hb_000062_400 |
0.1378068584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000086_210 |
0.1387954684 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_009079_050 |
0.1414012013 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 19 |
Hb_002129_020 |
0.1440387422 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 20 |
Hb_001049_150 |
0.1476215834 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |