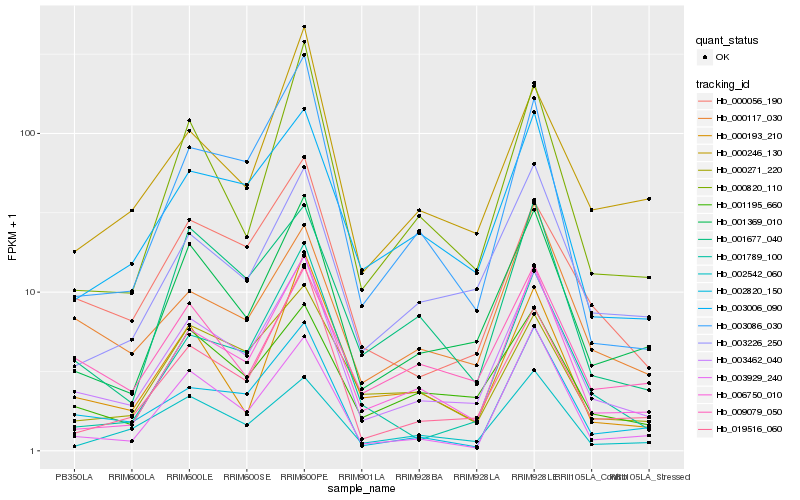
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003462_040 |
0.0 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_001369_010 |
0.0874079518 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 3 |
Hb_006750_010 |
0.1018085559 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 4 |
Hb_003006_090 |
0.1141164927 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 5 |
Hb_019516_060 |
0.1145425748 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 6 |
Hb_003226_250 |
0.1162290801 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 7 |
Hb_001195_660 |
0.1198727123 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 8 |
Hb_000056_190 |
0.1201640606 |
- |
- |
hypothetical protein [Ricinus communis] |
| 9 |
Hb_002820_150 |
0.1211664469 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000117_030 |
0.1322901277 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 [Jatropha curcas] |
| 11 |
Hb_001789_100 |
0.136758431 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001677_040 |
0.1369898864 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003929_240 |
0.1370935997 |
- |
- |
PREDICTED: uncharacterized protein LOC105643616 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002542_060 |
0.1377435265 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 15 |
Hb_000820_110 |
0.1385881554 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 16 |
Hb_000271_220 |
0.1424593968 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 17 |
Hb_000193_210 |
0.1425551125 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |
| 18 |
Hb_003086_030 |
0.143156566 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 19 |
Hb_009079_050 |
0.143417279 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 20 |
Hb_000246_130 |
0.1453920943 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |