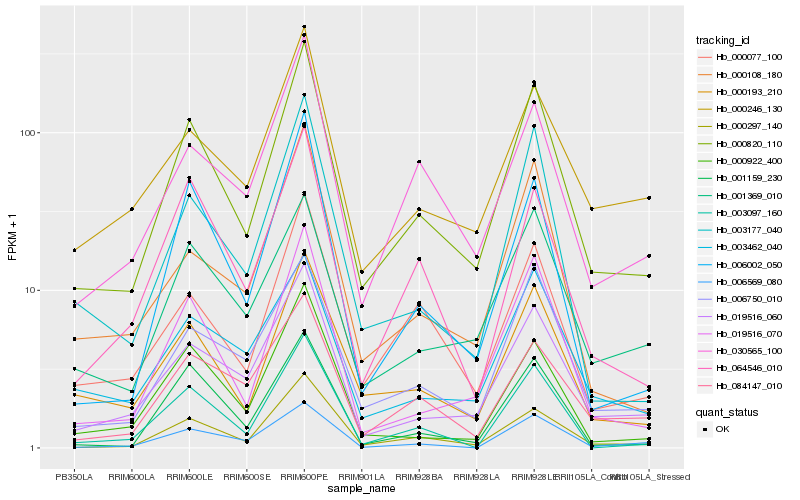
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000820_110 |
0.0 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 2 |
Hb_000297_140 |
0.0848614059 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 3 |
Hb_000193_210 |
0.0929900205 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |
| 4 |
Hb_019516_070 |
0.0955910803 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 5 |
Hb_019516_060 |
0.1040963776 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 6 |
Hb_006750_010 |
0.1160244329 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 7 |
Hb_000077_100 |
0.1197461654 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003177_040 |
0.1205309859 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 9 |
Hb_006002_050 |
0.1235224424 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 10 |
Hb_001369_010 |
0.1281798852 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 11 |
Hb_030565_100 |
0.1301710324 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000108_180 |
0.1301841589 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 13 |
Hb_000922_400 |
0.1307534471 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 14 |
Hb_003097_160 |
0.1323249892 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein sel-10 [Jatropha curcas] |
| 15 |
Hb_001159_230 |
0.1333535459 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 16 |
Hb_064546_010 |
0.1338658895 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 17 |
Hb_000246_130 |
0.1350603437 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 18 |
Hb_084147_010 |
0.1366343288 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_006569_080 |
0.1385489196 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_024559 [Vitis vinifera] |
| 20 |
Hb_003462_040 |
0.1385881554 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |