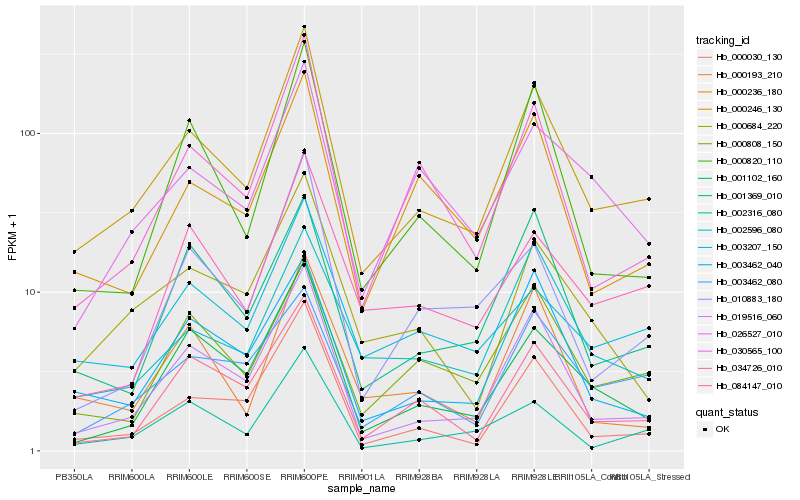
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000246_130 |
0.0 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 2 |
Hb_019516_060 |
0.0944647736 |
- |
- |
PREDICTED: uncharacterized protein LOC105634259 [Jatropha curcas] |
| 3 |
Hb_000030_130 |
0.1216422432 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_001102_160 |
0.1274140677 |
- |
- |
PREDICTED: galacturonokinase [Jatropha curcas] |
| 5 |
Hb_030565_100 |
0.129260051 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 6 |
Hb_002316_080 |
0.1330201765 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |
| 7 |
Hb_084147_010 |
0.1334882449 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_000820_110 |
0.1350603437 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 9 |
Hb_000684_220 |
0.1412589206 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000236_180 |
0.1424715773 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 11 |
Hb_003207_150 |
0.1426651929 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 12 |
Hb_000808_150 |
0.1439279687 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 13 |
Hb_003462_040 |
0.1453920943 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_010883_180 |
0.1469942447 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 15 |
Hb_003462_080 |
0.1470201988 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_026527_010 |
0.1477307767 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 17 |
Hb_002596_080 |
0.1500418164 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 18 |
Hb_034726_010 |
0.1507212603 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X2 [Jatropha curcas] |
| 19 |
Hb_001369_010 |
0.1527915863 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 20 |
Hb_000193_210 |
0.1527994249 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |