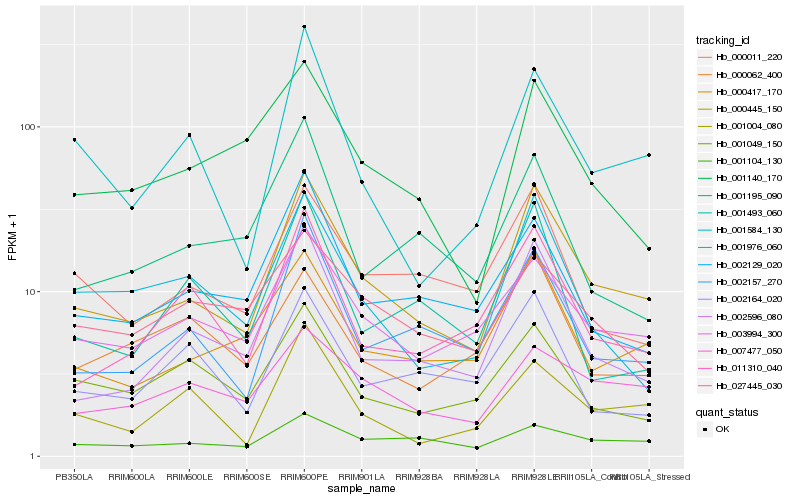
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_150 |
0.0 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 2 |
Hb_002596_080 |
0.1255765399 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 3 |
Hb_000417_170 |
0.1294774757 |
- |
- |
PREDICTED: uncharacterized protein LOC105649689 [Jatropha curcas] |
| 4 |
Hb_003994_300 |
0.1342627036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002157_270 |
0.1352116594 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 6 |
Hb_001584_130 |
0.1532966499 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 7 |
Hb_011310_040 |
0.153351119 |
transcription factor |
TF Family: bZIP |
transcription factor hy5, putative [Ricinus communis] |
| 8 |
Hb_001976_060 |
0.1574137182 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000011_220 |
0.1578934587 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 10 |
Hb_001004_080 |
0.1579908415 |
- |
- |
hypothetical protein POPTR_0014s14130g [Populus trichocarpa] |
| 11 |
Hb_002129_020 |
0.158039416 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 12 |
Hb_001140_170 |
0.1594545549 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 13 |
Hb_007477_050 |
0.1617000354 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001104_130 |
0.1711130941 |
- |
- |
C-14 sterol reductase, putative [Ricinus communis] |
| 15 |
Hb_001049_150 |
0.1722445723 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 16 |
Hb_001493_060 |
0.1729899531 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 17 |
Hb_001195_090 |
0.1739710775 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 18 |
Hb_002164_020 |
0.1748478017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000062_400 |
0.1770003179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_027445_030 |
0.179033087 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |