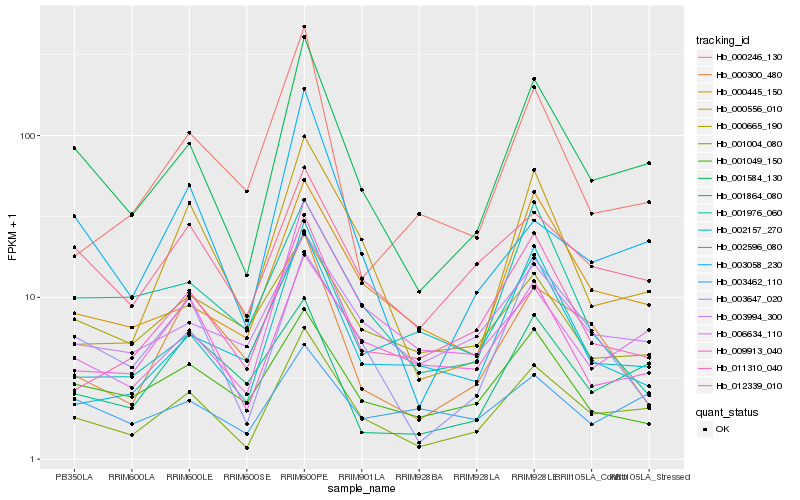
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001584_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 2 |
Hb_001004_080 |
0.0725473628 |
- |
- |
hypothetical protein POPTR_0014s14130g [Populus trichocarpa] |
| 3 |
Hb_000445_150 |
0.1532966499 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 4 |
Hb_003647_020 |
0.1681918853 |
- |
- |
hypothetical protein JCGZ_25095 [Jatropha curcas] |
| 5 |
Hb_012339_010 |
0.1701909052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001049_150 |
0.1788042152 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 7 |
Hb_003994_300 |
0.1808212163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002157_270 |
0.1844806939 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 9 |
Hb_011310_040 |
0.1874968395 |
transcription factor |
TF Family: bZIP |
transcription factor hy5, putative [Ricinus communis] |
| 10 |
Hb_003058_230 |
0.1894745826 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 11 |
Hb_000556_010 |
0.1896730428 |
- |
- |
glycerate dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_009913_040 |
0.1908049155 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 13 |
Hb_001976_060 |
0.1925347447 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006634_110 |
0.1934102189 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 15 |
Hb_001864_080 |
0.1954446149 |
- |
- |
PREDICTED: uncharacterized protein LOC105649405 [Jatropha curcas] |
| 16 |
Hb_003462_110 |
0.2022388834 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 17 |
Hb_000300_480 |
0.2025664589 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 18 |
Hb_000246_130 |
0.2040769344 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 19 |
Hb_002596_080 |
0.204206517 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 20 |
Hb_000665_190 |
0.2052459168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |