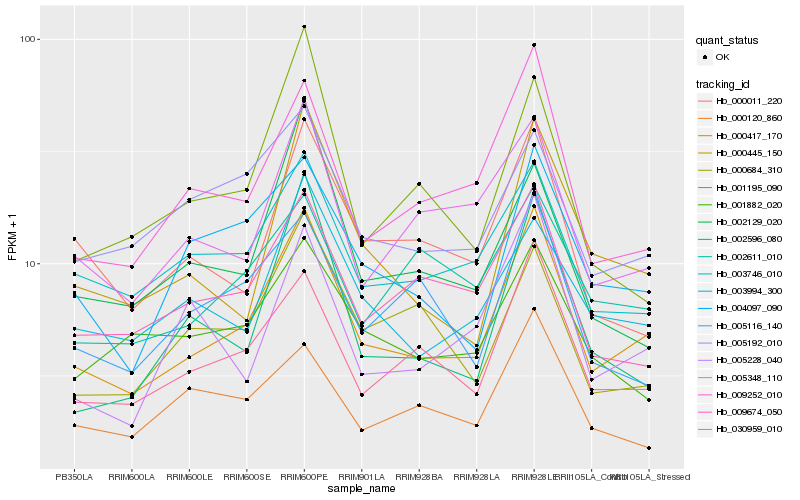
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649689 [Jatropha curcas] |
| 2 |
Hb_030959_010 |
0.1044115017 |
- |
- |
cullin, putative [Ricinus communis] |
| 3 |
Hb_009252_010 |
0.1139252855 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 4 |
Hb_003746_010 |
0.1264048258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004097_090 |
0.1292642242 |
- |
- |
PREDICTED: uncharacterized protein LOC105634517 [Jatropha curcas] |
| 6 |
Hb_000445_150 |
0.1294774757 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 7 |
Hb_005348_110 |
0.1305541162 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 8 |
Hb_009674_050 |
0.1320807307 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 9 |
Hb_003994_300 |
0.1323601029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000011_220 |
0.1371734448 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 11 |
Hb_002596_080 |
0.1428226386 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g04160 [Jatropha curcas] |
| 12 |
Hb_005192_010 |
0.1456076826 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 13 |
Hb_005228_040 |
0.1464424092 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 14 |
Hb_005116_140 |
0.1469934807 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 15 |
Hb_000684_310 |
0.1477997187 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 16 |
Hb_002611_010 |
0.150129087 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_001882_020 |
0.1502459167 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 18 |
Hb_002129_020 |
0.1505201713 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 19 |
Hb_000120_860 |
0.1515798121 |
- |
- |
nucellin, putative [Ricinus communis] |
| 20 |
Hb_001195_090 |
0.1555526686 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |