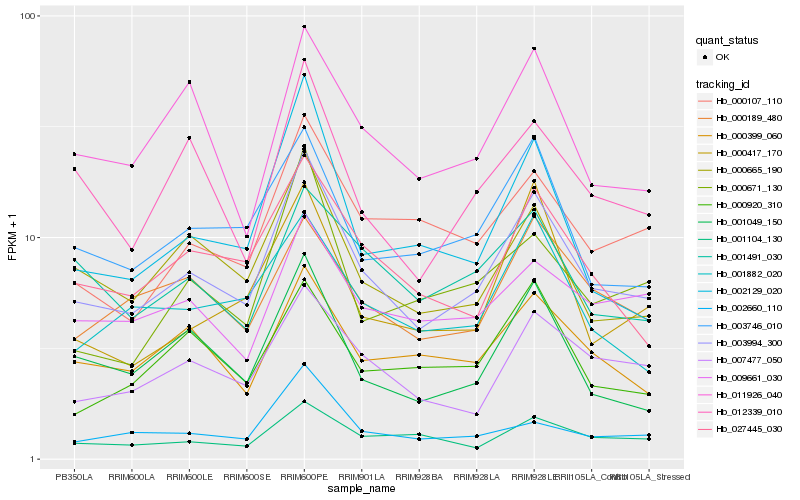
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003994_300 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000665_190 |
0.0997438272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_027445_030 |
0.1078553133 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_007477_050 |
0.1091228336 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003746_010 |
0.1123882307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001049_150 |
0.1164927687 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 7 |
Hb_000399_060 |
0.1177914451 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 8 |
Hb_012339_010 |
0.1195057895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000189_480 |
0.1195354085 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |
| 10 |
Hb_011926_040 |
0.1215398871 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000107_110 |
0.1218801234 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_001104_130 |
0.1219431984 |
- |
- |
C-14 sterol reductase, putative [Ricinus communis] |
| 13 |
Hb_001882_020 |
0.124150426 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 14 |
Hb_002660_110 |
0.1245330346 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009661_030 |
0.1253594313 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 16 |
Hb_002129_020 |
0.12602944 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 17 |
Hb_001491_030 |
0.1282802799 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000671_130 |
0.1304352265 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000920_310 |
0.1322520848 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 20 |
Hb_000417_170 |
0.1323601029 |
- |
- |
PREDICTED: uncharacterized protein LOC105649689 [Jatropha curcas] |