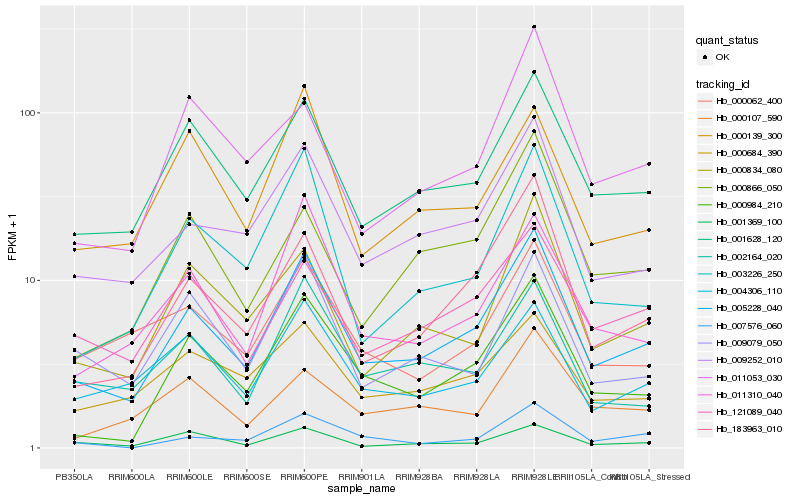
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005228_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 2 |
Hb_009252_010 |
0.1013067166 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 3 |
Hb_001628_120 |
0.1033893358 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 4 |
Hb_183963_010 |
0.1071002127 |
- |
- |
core region of GTP cyclohydrolase I family protein [Populus trichocarpa] |
| 5 |
Hb_000984_210 |
0.1100889678 |
- |
- |
PREDICTED: uncharacterized protein LOC104451821 [Eucalyptus grandis] |
| 6 |
Hb_003226_250 |
0.1231225889 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 7 |
Hb_000834_080 |
0.126986791 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 8 |
Hb_011310_040 |
0.1286619884 |
transcription factor |
TF Family: bZIP |
transcription factor hy5, putative [Ricinus communis] |
| 9 |
Hb_004306_110 |
0.1371633665 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 10 |
Hb_121089_040 |
0.1373618753 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 11 |
Hb_000684_390 |
0.138693158 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 12 |
Hb_002164_020 |
0.1388450052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000062_400 |
0.1412336988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001369_100 |
0.1415491421 |
- |
- |
hypothetical protein JCGZ_26280 [Jatropha curcas] |
| 15 |
Hb_009079_050 |
0.1422578877 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 16 |
Hb_007576_060 |
0.1430616494 |
- |
- |
- |
| 17 |
Hb_011053_030 |
0.1442025161 |
- |
- |
- |
| 18 |
Hb_000866_050 |
0.1442176257 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 19 |
Hb_000107_590 |
0.1446181691 |
- |
- |
PREDICTED: tricalbin-3 [Jatropha curcas] |
| 20 |
Hb_000139_300 |
0.1446943171 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |