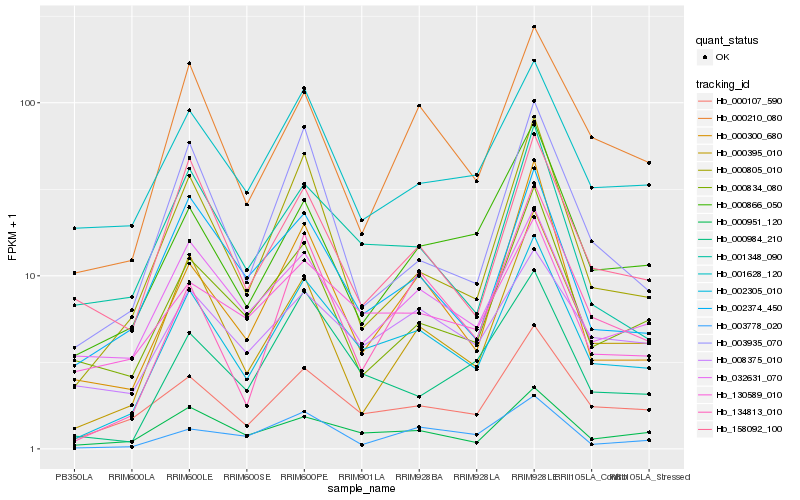
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002305_010 |
0.0 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000107_590 |
0.1085068594 |
- |
- |
PREDICTED: tricalbin-3 [Jatropha curcas] |
| 3 |
Hb_000951_120 |
0.1176399883 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 4 |
Hb_000805_010 |
0.1346857723 |
- |
- |
PREDICTED: uncharacterized protein LOC105629931 [Jatropha curcas] |
| 5 |
Hb_000210_080 |
0.1360472808 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000984_210 |
0.1389523592 |
- |
- |
PREDICTED: uncharacterized protein LOC104451821 [Eucalyptus grandis] |
| 7 |
Hb_002374_450 |
0.1399719323 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000395_010 |
0.1416730977 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 9 |
Hb_032631_070 |
0.1437063761 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_130589_010 |
0.1448947617 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 11 |
Hb_008375_010 |
0.1460557293 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 12 |
Hb_000866_050 |
0.1463666738 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 13 |
Hb_134813_010 |
0.1468616236 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 14 |
Hb_001628_120 |
0.147814978 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 15 |
Hb_001348_090 |
0.1493422415 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000300_680 |
0.1506555671 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003935_070 |
0.1509409901 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 18 |
Hb_158092_100 |
0.1512915082 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 19 |
Hb_000834_080 |
0.1519168275 |
- |
- |
PREDICTED: uncharacterized protein LOC105637718 [Jatropha curcas] |
| 20 |
Hb_003778_020 |
0.1543810021 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |